Meng Ye
The Influence of Text Variation on User Engagement in Cross-Platform Content Sharing
Apr 26, 2025Abstract:In today's cross-platform social media landscape, understanding factors that drive engagement for multimodal content, especially text paired with visuals, remains complex. This study investigates how rewriting Reddit post titles adapted from YouTube video titles affects user engagement. First, we build and analyze a large dataset of Reddit posts sharing YouTube videos, revealing that 21% of post titles are minimally modified. Statistical analysis demonstrates that title rewrites measurably improve engagement. Second, we design a controlled, multi-phase experiment to rigorously isolate the effects of textual variations by neutralizing confounding factors like video popularity, timing, and community norms. Comprehensive statistical tests reveal that effective title rewrites tend to feature emotional resonance, lexical richness, and alignment with community-specific norms. Lastly, pairwise ranking prediction experiments using a fine-tuned BERT classifier achieves 74% accuracy, significantly outperforming near-random baselines, including GPT-4o. These results validate that our controlled dataset effectively minimizes confounding effects, allowing advanced models to both learn and demonstrate the impact of textual features on engagement. By bridging quantitative rigor with qualitative insights, this study uncovers engagement dynamics and offers a robust framework for future cross-platform, multimodal content strategies.
Rate-My-LoRA: Efficient and Adaptive Federated Model Tuning for Cardiac MRI Segmentation
Jan 06, 2025Abstract:Cardiovascular disease (CVD) and cardiac dyssynchrony are major public health problems in the United States. Precise cardiac image segmentation is crucial for extracting quantitative measures that help categorize cardiac dyssynchrony. However, achieving high accuracy often depends on centralizing large datasets from different hospitals, which can be challenging due to privacy concerns. To solve this problem, Federated Learning (FL) is proposed to enable decentralized model training on such data without exchanging sensitive information. However, bandwidth limitations and data heterogeneity remain as significant challenges in conventional FL algorithms. In this paper, we propose a novel efficient and adaptive federate learning method for cardiac segmentation that improves model performance while reducing the bandwidth requirement. Our method leverages the low-rank adaptation (LoRA) to regularize model weight update and reduce communication overhead. We also propose a \mymethod{} aggregation technique to address data heterogeneity among clients. This technique adaptively penalizes the aggregated weights from different clients by comparing the validation accuracy in each client, allowing better generalization performance and fast local adaptation. In-client and cross-client evaluations on public cardiac MR datasets demonstrate the superiority of our method over other LoRA-based federate learning approaches.
VerSe: Integrating Multiple Queries as Prompts for Versatile Cardiac MRI Segmentation
Dec 20, 2024Abstract:Despite the advances in learning-based image segmentation approach, the accurate segmentation of cardiac structures from magnetic resonance imaging (MRI) remains a critical challenge. While existing automatic segmentation methods have shown promise, they still require extensive manual corrections of the segmentation results by human experts, particularly in complex regions such as the basal and apical parts of the heart. Recent efforts have been made on developing interactive image segmentation methods that enable human-in-the-loop learning. However, they are semi-automatic and inefficient, due to their reliance on click-based prompts, especially for 3D cardiac MRI volumes. To address these limitations, we propose VerSe, a Versatile Segmentation framework to unify automatic and interactive segmentation through mutiple queries. Our key innovation lies in the joint learning of object and click queries as prompts for a shared segmentation backbone. VerSe supports both fully automatic segmentation, through object queries, and interactive mask refinement, by providing click queries when needed. With the proposed integrated prompting scheme, VerSe demonstrates significant improvement in performance and efficiency over existing methods, on both cardiac MRI and out-of-distribution medical imaging datasets. The code is available at https://github.com/bangwayne/Verse.
Learning Volumetric Neural Deformable Models to Recover 3D Regional Heart Wall Motion from Multi-Planar Tagged MRI
Nov 21, 2024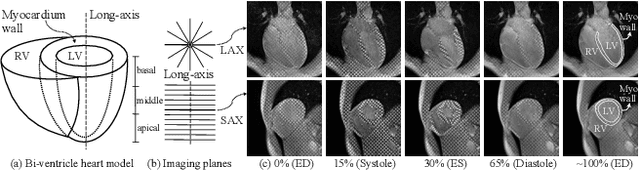
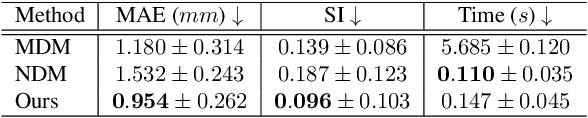
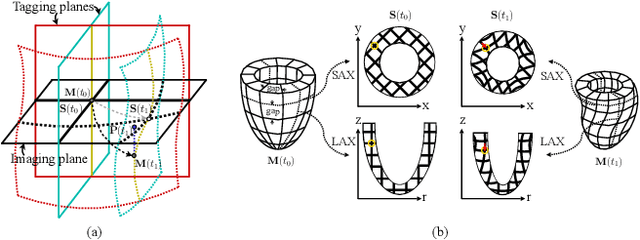
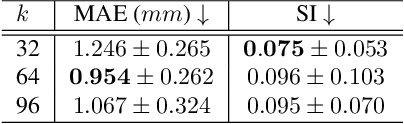
Abstract:Multi-planar tagged MRI is the gold standard for regional heart wall motion evaluation. However, accurate recovery of the 3D true heart wall motion from a set of 2D apparent motion cues is challenging, due to incomplete sampling of the true motion and difficulty in information fusion from apparent motion cues observed on multiple imaging planes. To solve these challenges, we introduce a novel class of volumetric neural deformable models ($\upsilon$NDMs). Our $\upsilon$NDMs represent heart wall geometry and motion through a set of low-dimensional global deformation parameter functions and a diffeomorphic point flow regularized local deformation field. To learn such global and local deformation for 2D apparent motion mapping to 3D true motion, we design a hybrid point transformer, which incorporates both point cross-attention and self-attention mechanisms. While use of point cross-attention can learn to fuse 2D apparent motion cues into material point true motion hints, point self-attention hierarchically organised as an encoder-decoder structure can further learn to refine these hints and map them into 3D true motion. We have performed experiments on a large cohort of synthetic 3D regional heart wall motion dataset. The results demonstrated the high accuracy of our method for the recovery of dense 3D true motion from sparse 2D apparent motion cues. Project page is at https://github.com/DeepTag/VolumetricNeuralDeformableModels.
Continuous Spatio-Temporal Memory Networks for 4D Cardiac Cine MRI Segmentation
Oct 30, 2024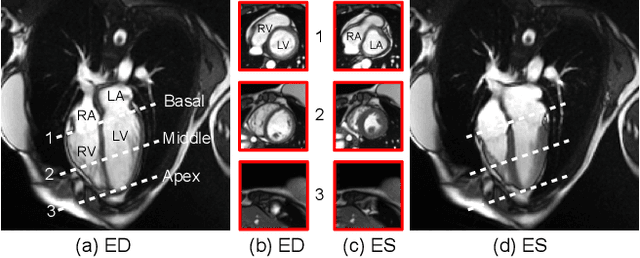
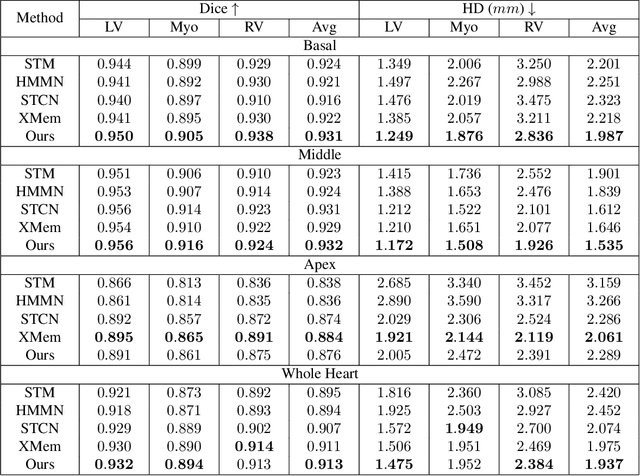
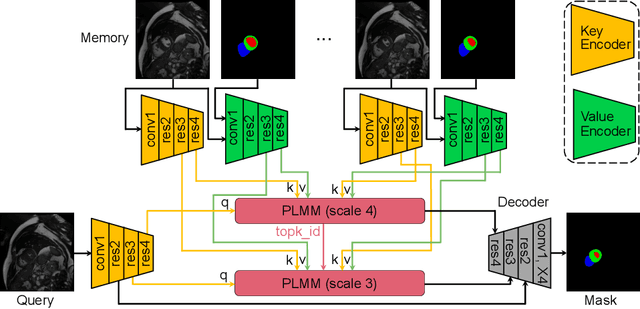
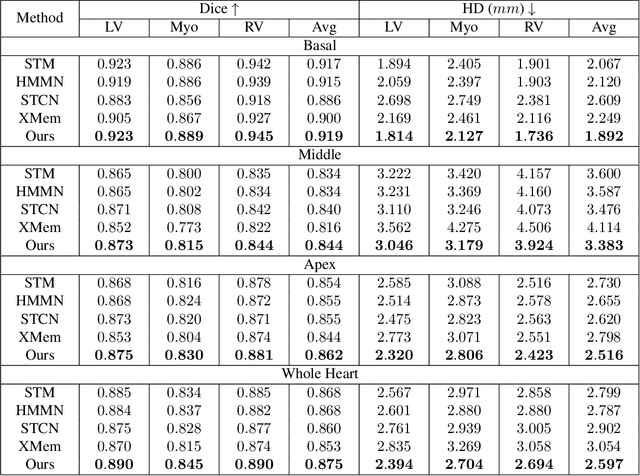
Abstract:Current cardiac cine magnetic resonance image (cMR) studies focus on the end diastole (ED) and end systole (ES) phases, while ignoring the abundant temporal information in the whole image sequence. This is because whole sequence segmentation is currently a tedious process and inaccurate. Conventional whole sequence segmentation approaches first estimate the motion field between frames, which is then used to propagate the mask along the temporal axis. However, the mask propagation results could be prone to error, especially for the basal and apex slices, where through-plane motion leads to significant morphology and structural change during the cardiac cycle. Inspired by recent advances in video object segmentation (VOS), based on spatio-temporal memory (STM) networks, we propose a continuous STM (CSTM) network for semi-supervised whole heart and whole sequence cMR segmentation. Our CSTM network takes full advantage of the spatial, scale, temporal and through-plane continuity prior of the underlying heart anatomy structures, to achieve accurate and fast 4D segmentation. Results of extensive experiments across multiple cMR datasets show that our method can improve the 4D cMR segmentation performance, especially for the hard-to-segment regions.
Empowering Interdisciplinary Insights with Dynamic Graph Embedding Trajectories
Jun 25, 2024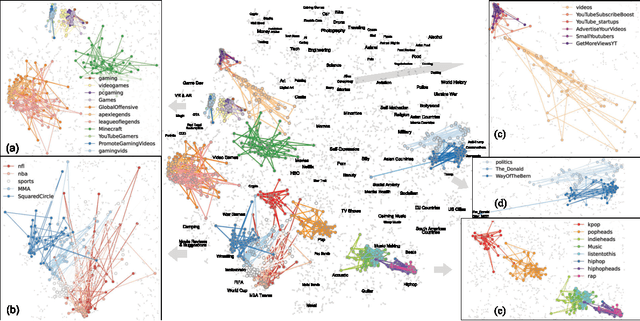
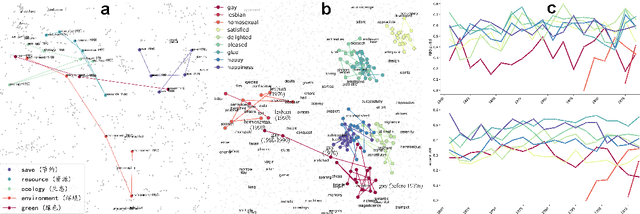
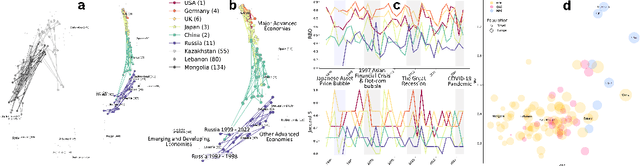
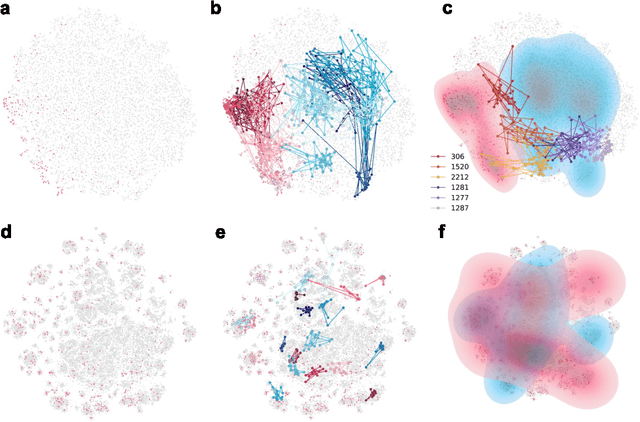
Abstract:We developed DyGETViz, a novel framework for effectively visualizing dynamic graphs (DGs) that are ubiquitous across diverse real-world systems. This framework leverages recent advancements in discrete-time dynamic graph (DTDG) models to adeptly handle the temporal dynamics inherent in dynamic graphs. DyGETViz effectively captures both micro- and macro-level structural shifts within these graphs, offering a robust method for representing complex and massive dynamic graphs. The application of DyGETViz extends to a diverse array of domains, including ethology, epidemiology, finance, genetics, linguistics, communication studies, social studies, and international relations. Through its implementation, DyGETViz has revealed or confirmed various critical insights. These include the diversity of content sharing patterns and the degree of specialization within online communities, the chronological evolution of lexicons across decades, and the distinct trajectories exhibited by aging-related and non-related genes. Importantly, DyGETViz enhances the accessibility of scientific findings to non-domain experts by simplifying the complexities of dynamic graphs. Our framework is released as an open-source Python package for use across diverse disciplines. Our work not only addresses the ongoing challenges in visualizing and analyzing DTDG models but also establishes a foundational framework for future investigations into dynamic graph representation and analysis across various disciplines.
A Video is Worth 10,000 Words: Training and Benchmarking with Diverse Captions for Better Long Video Retrieval
Nov 30, 2023



Abstract:Existing long video retrieval systems are trained and tested in the paragraph-to-video retrieval regime, where every long video is described by a single long paragraph. This neglects the richness and variety of possible valid descriptions of a video, which could be described in moment-by-moment detail, or in a single phrase summary, or anything in between. To provide a more thorough evaluation of the capabilities of long video retrieval systems, we propose a pipeline that leverages state-of-the-art large language models to carefully generate a diverse set of synthetic captions for long videos. We validate this pipeline's fidelity via rigorous human inspection. We then benchmark a representative set of video language models on these synthetic captions using a few long video datasets, showing that they struggle with the transformed data, especially the shortest captions. We also propose a lightweight fine-tuning method, where we use a contrastive loss to learn a hierarchical embedding loss based on the differing levels of information among the various captions. Our method improves performance both on the downstream paragraph-to-video retrieval task (+1.1% R@1 on ActivityNet), as well as for the various long video retrieval metrics we compute using our synthetic data (+3.6% R@1 for short descriptions on ActivityNet). For data access and other details, please refer to our project website at https://mgwillia.github.io/10k-words.
Fill the K-Space and Refine the Image: Prompting for Dynamic and Multi-Contrast MRI Reconstruction
Sep 25, 2023



Abstract:The key to dynamic or multi-contrast magnetic resonance imaging (MRI) reconstruction lies in exploring inter-frame or inter-contrast information. Currently, the unrolled model, an approach combining iterative MRI reconstruction steps with learnable neural network layers, stands as the best-performing method for MRI reconstruction. However, there are two main limitations to overcome: firstly, the unrolled model structure and GPU memory constraints restrict the capacity of each denoising block in the network, impeding the effective extraction of detailed features for reconstruction; secondly, the existing model lacks the flexibility to adapt to variations in the input, such as different contrasts, resolutions or views, necessitating the training of separate models for each input type, which is inefficient and may lead to insufficient reconstruction. In this paper, we propose a two-stage MRI reconstruction pipeline to address these limitations. The first stage involves filling the missing k-space data, which we approach as a physics-based reconstruction problem. We first propose a simple yet efficient baseline model, which utilizes adjacent frames/contrasts and channel attention to capture the inherent inter-frame/-contrast correlation. Then, we extend the baseline model to a prompt-based learning approach, PromptMR, for all-in-one MRI reconstruction from different views, contrasts, adjacent types, and acceleration factors. The second stage is to refine the reconstruction from the first stage, which we treat as a general video restoration problem to further fuse features from neighboring frames/contrasts in the image domain. Extensive experiments show that our proposed method significantly outperforms previous state-of-the-art accelerated MRI reconstruction methods.
DeFormer: Integrating Transformers with Deformable Models for 3D Shape Abstraction from a Single Image
Sep 22, 2023
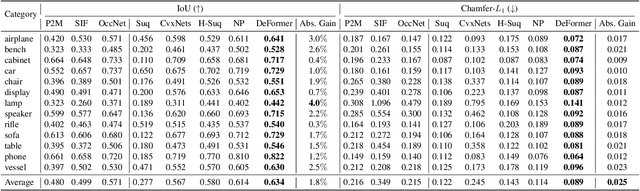
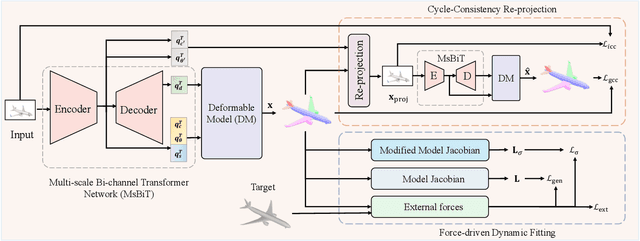
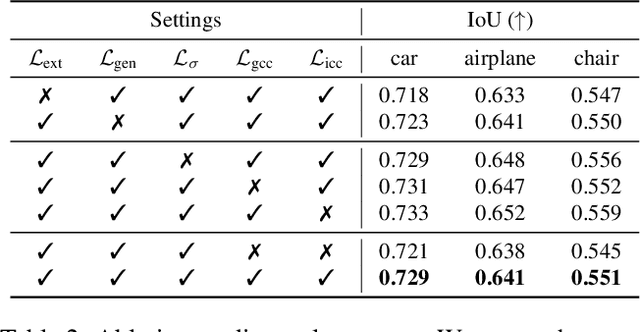
Abstract:Accurate 3D shape abstraction from a single 2D image is a long-standing problem in computer vision and graphics. By leveraging a set of primitives to represent the target shape, recent methods have achieved promising results. However, these methods either use a relatively large number of primitives or lack geometric flexibility due to the limited expressibility of the primitives. In this paper, we propose a novel bi-channel Transformer architecture, integrated with parameterized deformable models, termed DeFormer, to simultaneously estimate the global and local deformations of primitives. In this way, DeFormer can abstract complex object shapes while using a small number of primitives which offer a broader geometry coverage and finer details. Then, we introduce a force-driven dynamic fitting and a cycle-consistent re-projection loss to optimize the primitive parameters. Extensive experiments on ShapeNet across various settings show that DeFormer achieves better reconstruction accuracy over the state-of-the-art, and visualizes with consistent semantic correspondences for improved interpretability.
Neural Deformable Models for 3D Bi-Ventricular Heart Shape Reconstruction and Modeling from 2D Sparse Cardiac Magnetic Resonance Imaging
Jul 15, 2023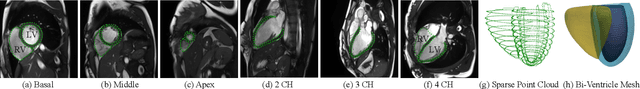
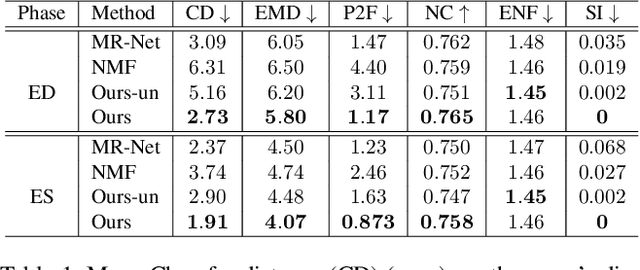
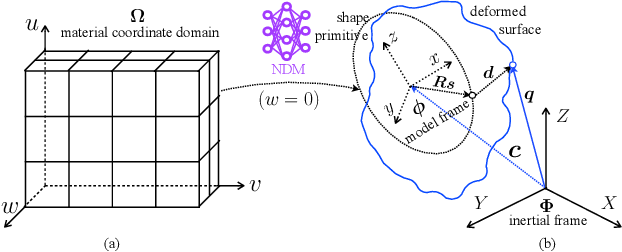

Abstract:We propose a novel neural deformable model (NDM) targeting at the reconstruction and modeling of 3D bi-ventricular shape of the heart from 2D sparse cardiac magnetic resonance (CMR) imaging data. We model the bi-ventricular shape using blended deformable superquadrics, which are parameterized by a set of geometric parameter functions and are capable of deforming globally and locally. While global geometric parameter functions and deformations capture gross shape features from visual data, local deformations, parameterized as neural diffeomorphic point flows, can be learned to recover the detailed heart shape.Different from iterative optimization methods used in conventional deformable model formulations, NDMs can be trained to learn such geometric parameter functions, global and local deformations from a shape distribution manifold. Our NDM can learn to densify a sparse cardiac point cloud with arbitrary scales and generate high-quality triangular meshes automatically. It also enables the implicit learning of dense correspondences among different heart shape instances for accurate cardiac shape registration. Furthermore, the parameters of NDM are intuitive, and can be used by a physician without sophisticated post-processing. Experimental results on a large CMR dataset demonstrate the improved performance of NDM over conventional methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge