Yunhe Gao
Atlas is Your Perfect Context: One-Shot Customization for Generalizable Foundational Medical Image Segmentation
Dec 20, 2025



Abstract:Accurate medical image segmentation is essential for clinical diagnosis and treatment planning. While recent interactive foundation models (e.g., nnInteractive) enhance generalization through large-scale multimodal pretraining, they still depend on precise prompts and often perform below expectations in contexts that are underrepresented in their training data. We present AtlasSegFM, an atlas-guided framework that customizes available foundation models to clinical contexts with a single annotated example. The core innovations are: 1) a pipeline that provides context-aware prompts for foundation models via registration between a context atlas and query images, and 2) a test-time adapter to fuse predictions from both atlas registration and the foundation model. Extensive experiments across public and in-house datasets spanning multiple modalities and organs demonstrate that AtlasSegFM consistently improves segmentation, particularly for small, delicate structures. AtlasSegFM provides a lightweight, deployable solution one-shot customization of foundation models in real-world clinical workflows. The code will be made publicly available.
Anatomy-VLM: A Fine-grained Vision-Language Model for Medical Interpretation
Nov 11, 2025Abstract:Accurate disease interpretation from radiology remains challenging due to imaging heterogeneity. Achieving expert-level diagnostic decisions requires integration of subtle image features with clinical knowledge. Yet major vision-language models (VLMs) treat images as holistic entities and overlook fine-grained image details that are vital for disease diagnosis. Clinicians analyze images by utilizing their prior medical knowledge and identify anatomical structures as important region of interests (ROIs). Inspired from this human-centric workflow, we introduce Anatomy-VLM, a fine-grained, vision-language model that incorporates multi-scale information. First, we design a model encoder to localize key anatomical features from entire medical images. Second, these regions are enriched with structured knowledge for contextually-aware interpretation. Finally, the model encoder aligns multi-scale medical information to generate clinically-interpretable disease prediction. Anatomy-VLM achieves outstanding performance on both in- and out-of-distribution datasets. We also validate the performance of Anatomy-VLM on downstream image segmentation tasks, suggesting that its fine-grained alignment captures anatomical and pathology-related knowledge. Furthermore, the Anatomy-VLM's encoder facilitates zero-shot anatomy-wise interpretation, providing its strong expert-level clinical interpretation capabilities.
Show and Segment: Universal Medical Image Segmentation via In-Context Learning
Mar 25, 2025Abstract:Medical image segmentation remains challenging due to the vast diversity of anatomical structures, imaging modalities, and segmentation tasks. While deep learning has made significant advances, current approaches struggle to generalize as they require task-specific training or fine-tuning on unseen classes. We present Iris, a novel In-context Reference Image guided Segmentation framework that enables flexible adaptation to novel tasks through the use of reference examples without fine-tuning. At its core, Iris features a lightweight context task encoding module that distills task-specific information from reference context image-label pairs. This rich context embedding information is used to guide the segmentation of target objects. By decoupling task encoding from inference, Iris supports diverse strategies from one-shot inference and context example ensemble to object-level context example retrieval and in-context tuning. Through comprehensive evaluation across twelve datasets, we demonstrate that Iris performs strongly compared to task-specific models on in-distribution tasks. On seven held-out datasets, Iris shows superior generalization to out-of-distribution data and unseen classes. Further, Iris's task encoding module can automatically discover anatomical relationships across datasets and modalities, offering insights into medical objects without explicit anatomical supervision.
The Hidden Life of Tokens: Reducing Hallucination of Large Vision-Language Models via Visual Information Steering
Feb 05, 2025



Abstract:Large Vision-Language Models (LVLMs) can reason effectively over both textual and visual inputs, but they tend to hallucinate syntactically coherent yet visually ungrounded contents. In this paper, we investigate the internal dynamics of hallucination by examining the tokens logits rankings throughout the generation process, revealing three key patterns in how LVLMs process information: (1) gradual visual information loss -- visually grounded tokens gradually become less favored throughout generation, and (2) early excitation -- semantically meaningful tokens achieve peak activation in the layers earlier than the final layer. (3) hidden genuine information -- visually grounded tokens though not being eventually decided still retain relatively high rankings at inference. Based on these insights, we propose VISTA (Visual Information Steering with Token-logit Augmentation), a training-free inference-time intervention framework that reduces hallucination while promoting genuine information. VISTA works by combining two complementary approaches: reinforcing visual information in activation space and leveraging early layer activations to promote semantically meaningful decoding. Compared to existing methods, VISTA requires no external supervision and is applicable to various decoding strategies. Extensive experiments show that VISTA on average reduces hallucination by abount 40% on evaluated open-ended generation task, and it consistently outperforms existing methods on four benchmarks across four architectures under three decoding strategies.
RadAlign: Advancing Radiology Report Generation with Vision-Language Concept Alignment
Jan 13, 2025Abstract:Automated chest radiographs interpretation requires both accurate disease classification and detailed radiology report generation, presenting a significant challenge in the clinical workflow. Current approaches either focus on classification accuracy at the expense of interpretability or generate detailed but potentially unreliable reports through image captioning techniques. In this study, we present RadAlign, a novel framework that combines the predictive accuracy of vision-language models (VLMs) with the reasoning capabilities of large language models (LLMs). Inspired by the radiologist's workflow, RadAlign first employs a specialized VLM to align visual features with key medical concepts, achieving superior disease classification with an average AUC of 0.885 across multiple diseases. These recognized medical conditions, represented as text-based concepts in the aligned visual-language space, are then used to prompt LLM-based report generation. Enhanced by a retrieval-augmented generation mechanism that grounds outputs in similar historical cases, RadAlign delivers superior report quality with a GREEN score of 0.678, outperforming state-of-the-art methods' 0.634. Our framework maintains strong clinical interpretability while reducing hallucinations, advancing automated medical imaging and report analysis through integrated predictive and generative AI. Code is available at https://github.com/difeigu/RadAlign.
VerSe: Integrating Multiple Queries as Prompts for Versatile Cardiac MRI Segmentation
Dec 20, 2024Abstract:Despite the advances in learning-based image segmentation approach, the accurate segmentation of cardiac structures from magnetic resonance imaging (MRI) remains a critical challenge. While existing automatic segmentation methods have shown promise, they still require extensive manual corrections of the segmentation results by human experts, particularly in complex regions such as the basal and apical parts of the heart. Recent efforts have been made on developing interactive image segmentation methods that enable human-in-the-loop learning. However, they are semi-automatic and inefficient, due to their reliance on click-based prompts, especially for 3D cardiac MRI volumes. To address these limitations, we propose VerSe, a Versatile Segmentation framework to unify automatic and interactive segmentation through mutiple queries. Our key innovation lies in the joint learning of object and click queries as prompts for a shared segmentation backbone. VerSe supports both fully automatic segmentation, through object queries, and interactive mask refinement, by providing click queries when needed. With the proposed integrated prompting scheme, VerSe demonstrates significant improvement in performance and efficiency over existing methods, on both cardiac MRI and out-of-distribution medical imaging datasets. The code is available at https://github.com/bangwayne/Verse.
Aligning Human Knowledge with Visual Concepts Towards Explainable Medical Image Classification
Jun 08, 2024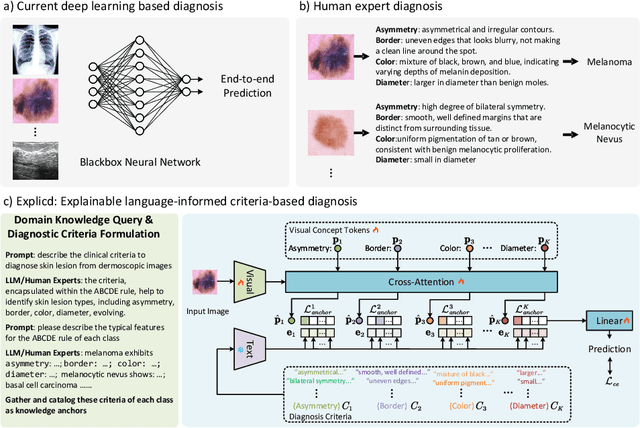
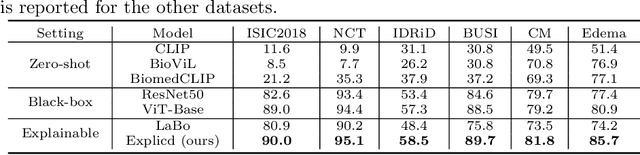
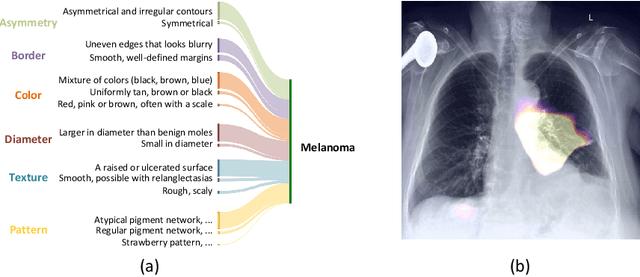
Abstract:Although explainability is essential in the clinical diagnosis, most deep learning models still function as black boxes without elucidating their decision-making process. In this study, we investigate the explainable model development that can mimic the decision-making process of human experts by fusing the domain knowledge of explicit diagnostic criteria. We introduce a simple yet effective framework, Explicd, towards Explainable language-informed criteria-based diagnosis. Explicd initiates its process by querying domain knowledge from either large language models (LLMs) or human experts to establish diagnostic criteria across various concept axes (e.g., color, shape, texture, or specific patterns of diseases). By leveraging a pretrained vision-language model, Explicd injects these criteria into the embedding space as knowledge anchors, thereby facilitating the learning of corresponding visual concepts within medical images. The final diagnostic outcome is determined based on the similarity scores between the encoded visual concepts and the textual criteria embeddings. Through extensive evaluation of five medical image classification benchmarks, Explicd has demonstrated its inherent explainability and extends to improve classification performance compared to traditional black-box models.
Implicit In-context Learning
May 23, 2024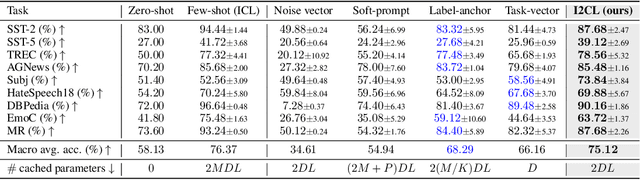
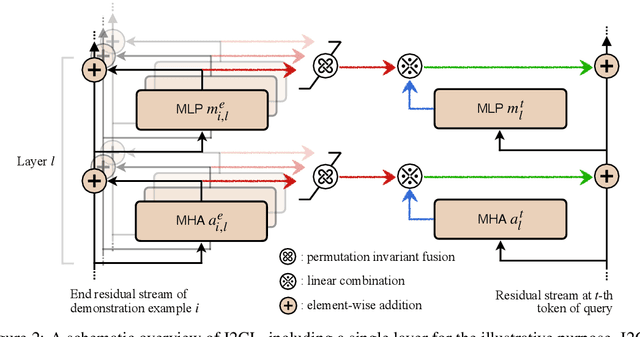
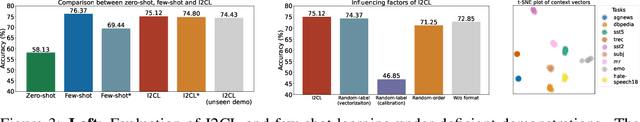
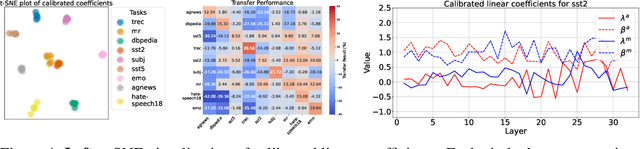
Abstract:In-context Learning (ICL) empowers large language models (LLMs) to adapt to unseen tasks during inference by prefixing a few demonstration examples prior to test queries. Despite its versatility, ICL incurs substantial computational and memory overheads compared to zero-shot learning and is susceptible to the selection and order of demonstration examples. In this work, we introduce Implicit In-context Learning (I2CL), an innovative paradigm that addresses the challenges associated with traditional ICL by absorbing demonstration examples within the activation space. I2CL first generates a condensed vector representation, namely a context vector, from the demonstration examples. It then integrates the context vector during inference by injecting a linear combination of the context vector and query activations into the model's residual streams. Empirical evaluation on nine real-world tasks across three model architectures demonstrates that I2CL achieves few-shot performance with zero-shot cost and exhibits robustness against the variation of demonstration examples. Furthermore, I2CL facilitates a novel representation of "task-ids", enhancing task similarity detection and enabling effective transfer learning. We provide a comprehensive analysis of I2CL, offering deeper insights into its mechanisms and broader implications for ICL. The source code is available at: https://github.com/LzVv123456/I2CL.
Deep Deformable Models: Learning 3D Shape Abstractions with Part Consistency
Sep 02, 2023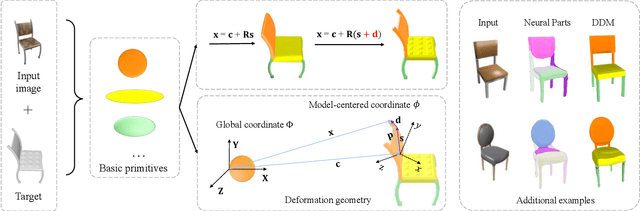
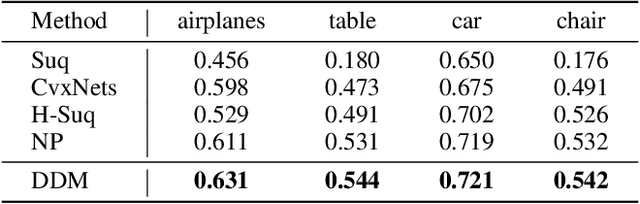
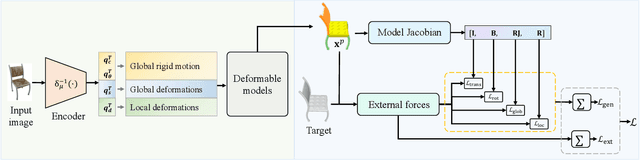
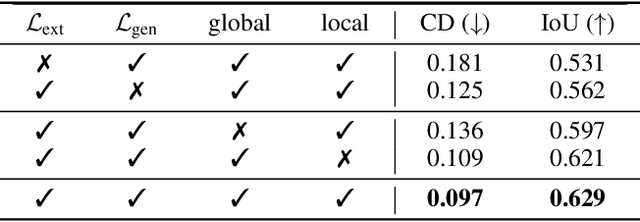
Abstract:The task of shape abstraction with semantic part consistency is challenging due to the complex geometries of natural objects. Recent methods learn to represent an object shape using a set of simple primitives to fit the target. \textcolor{black}{However, in these methods, the primitives used do not always correspond to real parts or lack geometric flexibility for semantic interpretation.} In this paper, we investigate salient and efficient primitive descriptors for accurate shape abstractions, and propose \textit{Deep Deformable Models (DDMs)}. DDM employs global deformations and diffeomorphic local deformations. These properties enable DDM to abstract complex object shapes with significantly fewer primitives that offer broader geometry coverage and finer details. DDM is also capable of learning part-level semantic correspondences due to the differentiable and invertible properties of our primitive deformation. Moreover, DDM learning formulation is based on dynamic and kinematic modeling, which enables joint regularization of each sub-transformation during primitive fitting. Extensive experiments on \textit{ShapeNet} demonstrate that DDM outperforms the state-of-the-art in terms of reconstruction and part consistency by a notable margin.
Training Like a Medical Resident: Universal Medical Image Segmentation via Context Prior Learning
Jun 06, 2023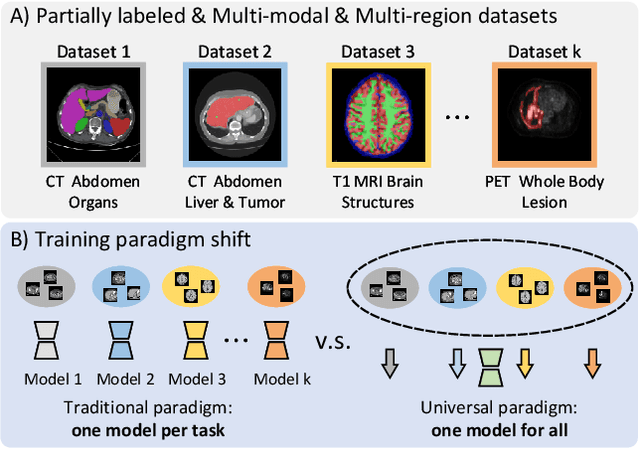
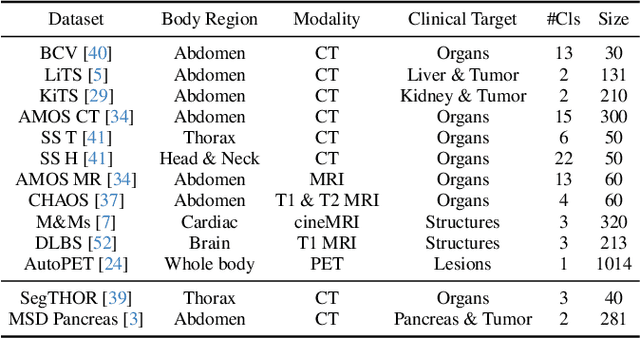
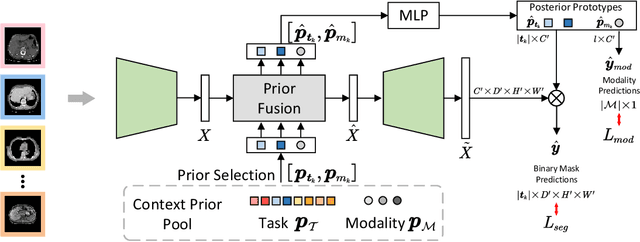
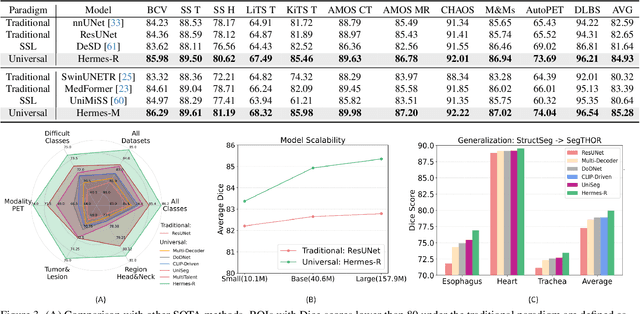
Abstract:A major enduring focus of clinical workflows is disease analytics and diagnosis, leading to medical imaging datasets where the modalities and annotations are strongly tied to specific clinical objectives. To date, building task-specific segmentation models is intuitive yet a restrictive approach, lacking insights gained from widespread imaging cohorts. Inspired by the training of medical residents, we explore universal medical image segmentation, whose goal is to learn from diverse medical imaging sources covering a range of clinical targets, body regions, and image modalities. Following this paradigm, we propose Hermes, a context prior learning approach that addresses the challenges related to the heterogeneity on data, modality, and annotations in the proposed universal paradigm. In a collection of seven diverse datasets, we demonstrate the appealing merits of the universal paradigm over the traditional task-specific training paradigm. By leveraging the synergy among various tasks, Hermes shows superior performance and model scalability. Our in-depth investigation on two additional datasets reveals Hermes' strong capabilities for transfer learning, incremental learning, and generalization to different downstream tasks. The code is available: https://github.com/yhygao/universal-medical-image-segmentation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge