Mu Zhou
Anatomy-VLM: A Fine-grained Vision-Language Model for Medical Interpretation
Nov 11, 2025Abstract:Accurate disease interpretation from radiology remains challenging due to imaging heterogeneity. Achieving expert-level diagnostic decisions requires integration of subtle image features with clinical knowledge. Yet major vision-language models (VLMs) treat images as holistic entities and overlook fine-grained image details that are vital for disease diagnosis. Clinicians analyze images by utilizing their prior medical knowledge and identify anatomical structures as important region of interests (ROIs). Inspired from this human-centric workflow, we introduce Anatomy-VLM, a fine-grained, vision-language model that incorporates multi-scale information. First, we design a model encoder to localize key anatomical features from entire medical images. Second, these regions are enriched with structured knowledge for contextually-aware interpretation. Finally, the model encoder aligns multi-scale medical information to generate clinically-interpretable disease prediction. Anatomy-VLM achieves outstanding performance on both in- and out-of-distribution datasets. We also validate the performance of Anatomy-VLM on downstream image segmentation tasks, suggesting that its fine-grained alignment captures anatomical and pathology-related knowledge. Furthermore, the Anatomy-VLM's encoder facilitates zero-shot anatomy-wise interpretation, providing its strong expert-level clinical interpretation capabilities.
Show and Segment: Universal Medical Image Segmentation via In-Context Learning
Mar 25, 2025Abstract:Medical image segmentation remains challenging due to the vast diversity of anatomical structures, imaging modalities, and segmentation tasks. While deep learning has made significant advances, current approaches struggle to generalize as they require task-specific training or fine-tuning on unseen classes. We present Iris, a novel In-context Reference Image guided Segmentation framework that enables flexible adaptation to novel tasks through the use of reference examples without fine-tuning. At its core, Iris features a lightweight context task encoding module that distills task-specific information from reference context image-label pairs. This rich context embedding information is used to guide the segmentation of target objects. By decoupling task encoding from inference, Iris supports diverse strategies from one-shot inference and context example ensemble to object-level context example retrieval and in-context tuning. Through comprehensive evaluation across twelve datasets, we demonstrate that Iris performs strongly compared to task-specific models on in-distribution tasks. On seven held-out datasets, Iris shows superior generalization to out-of-distribution data and unseen classes. Further, Iris's task encoding module can automatically discover anatomical relationships across datasets and modalities, offering insights into medical objects without explicit anatomical supervision.
MedForge: Building Medical Foundation Models Like Open Source Software Development
Feb 22, 2025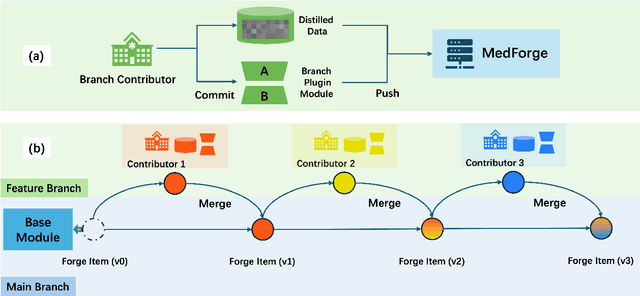
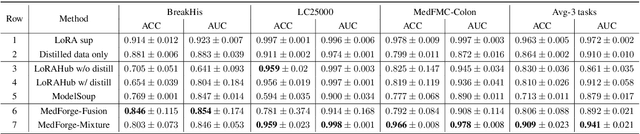
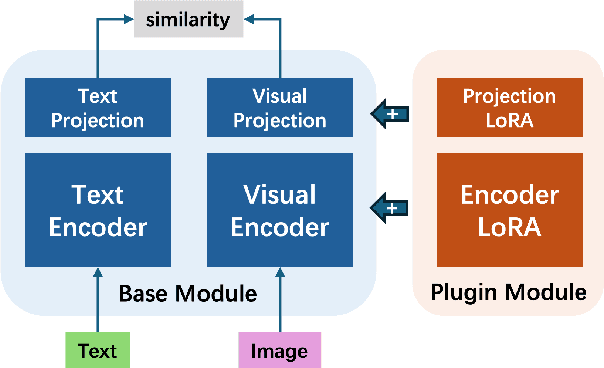
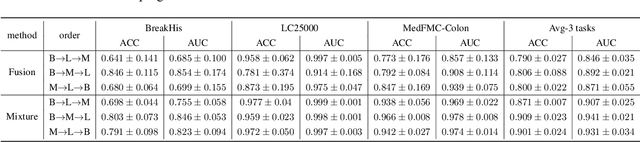
Abstract:Foundational models (FMs) have made significant strides in the healthcare domain. Yet the data silo challenge and privacy concern remain in healthcare systems, hindering safe medical data sharing and collaborative model development among institutions. The collection and curation of scalable clinical datasets increasingly become the bottleneck for training strong FMs. In this study, we propose Medical Foundation Models Merging (MedForge), a cooperative framework enabling a community-driven medical foundation model development, meanwhile preventing the information leakage of raw patient data and mitigating synchronization model development issues across clinical institutions. MedForge offers a bottom-up model construction mechanism by flexibly merging task-specific Low-Rank Adaptation (LoRA) modules, which can adapt to downstream tasks while retaining original model parameters. Through an asynchronous LoRA module integration scheme, the resulting composite model can progressively enhance its comprehensive performance on various clinical tasks. MedForge shows strong performance on multiple clinical datasets (e.g., breast cancer, lung cancer, and colon cancer) collected from different institutions. Our major findings highlight the value of collaborative foundation models in advancing multi-center clinical collaboration effectively and cohesively. Our code is publicly available at https://github.com/TanZheling/MedForge.
RadAlign: Advancing Radiology Report Generation with Vision-Language Concept Alignment
Jan 13, 2025Abstract:Automated chest radiographs interpretation requires both accurate disease classification and detailed radiology report generation, presenting a significant challenge in the clinical workflow. Current approaches either focus on classification accuracy at the expense of interpretability or generate detailed but potentially unreliable reports through image captioning techniques. In this study, we present RadAlign, a novel framework that combines the predictive accuracy of vision-language models (VLMs) with the reasoning capabilities of large language models (LLMs). Inspired by the radiologist's workflow, RadAlign first employs a specialized VLM to align visual features with key medical concepts, achieving superior disease classification with an average AUC of 0.885 across multiple diseases. These recognized medical conditions, represented as text-based concepts in the aligned visual-language space, are then used to prompt LLM-based report generation. Enhanced by a retrieval-augmented generation mechanism that grounds outputs in similar historical cases, RadAlign delivers superior report quality with a GREEN score of 0.678, outperforming state-of-the-art methods' 0.634. Our framework maintains strong clinical interpretability while reducing hallucinations, advancing automated medical imaging and report analysis through integrated predictive and generative AI. Code is available at https://github.com/difeigu/RadAlign.
Cost-effective Instruction Learning for Pathology Vision and Language Analysis
Jul 25, 2024
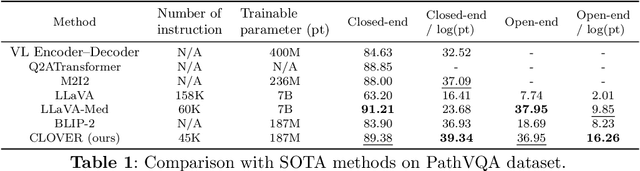
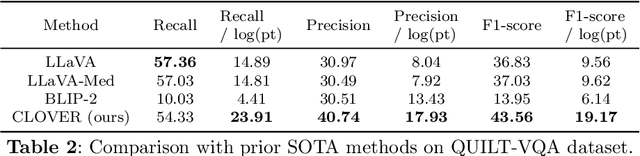

Abstract:The advent of vision-language models fosters the interactive conversations between AI-enabled models and humans. Yet applying these models into clinics must deal with daunting challenges around large-scale training data, financial, and computational resources. Here we propose a cost-effective instruction learning framework for conversational pathology named as CLOVER. CLOVER only trains a lightweight module and uses instruction tuning while freezing the parameters of the large language model. Instead of using costly GPT-4, we propose well-designed prompts on GPT-3.5 for building generation-based instructions, emphasizing the utility of pathological knowledge derived from the Internet source. To augment the use of instructions, we construct a high-quality set of template-based instructions in the context of digital pathology. From two benchmark datasets, our findings reveal the strength of hybrid-form instructions in the visual question-answer in pathology. Extensive results show the cost-effectiveness of CLOVER in answering both open-ended and closed-ended questions, where CLOVER outperforms strong baselines that possess 37 times more training parameters and use instruction data generated from GPT-4. Through the instruction tuning, CLOVER exhibits robustness of few-shot learning in the external clinical dataset. These findings demonstrate that cost-effective modeling of CLOVER could accelerate the adoption of rapid conversational applications in the landscape of digital pathology.
Aligning Human Knowledge with Visual Concepts Towards Explainable Medical Image Classification
Jun 08, 2024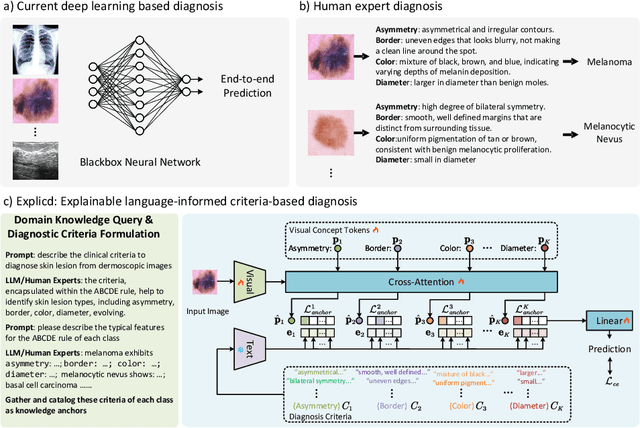
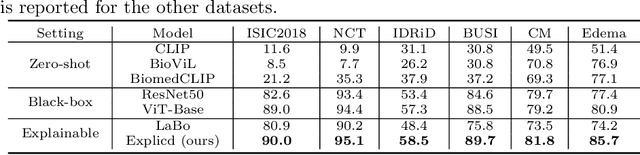
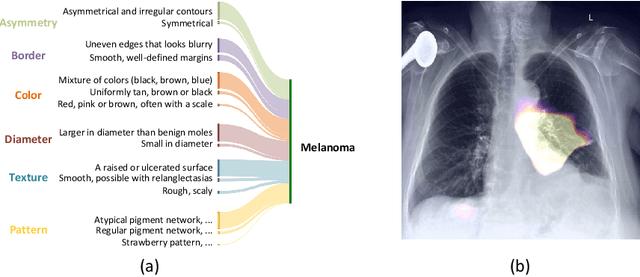
Abstract:Although explainability is essential in the clinical diagnosis, most deep learning models still function as black boxes without elucidating their decision-making process. In this study, we investigate the explainable model development that can mimic the decision-making process of human experts by fusing the domain knowledge of explicit diagnostic criteria. We introduce a simple yet effective framework, Explicd, towards Explainable language-informed criteria-based diagnosis. Explicd initiates its process by querying domain knowledge from either large language models (LLMs) or human experts to establish diagnostic criteria across various concept axes (e.g., color, shape, texture, or specific patterns of diseases). By leveraging a pretrained vision-language model, Explicd injects these criteria into the embedding space as knowledge anchors, thereby facilitating the learning of corresponding visual concepts within medical images. The final diagnostic outcome is determined based on the similarity scores between the encoded visual concepts and the textual criteria embeddings. Through extensive evaluation of five medical image classification benchmarks, Explicd has demonstrated its inherent explainability and extends to improve classification performance compared to traditional black-box models.
Data-Centric Foundation Models in Computational Healthcare: A Survey
Jan 04, 2024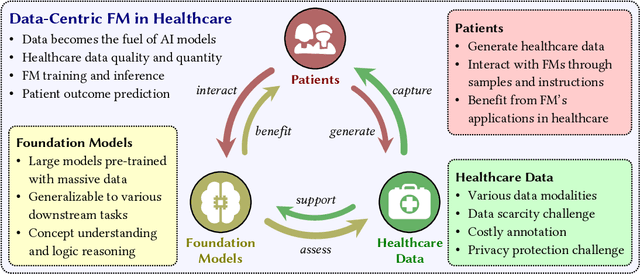
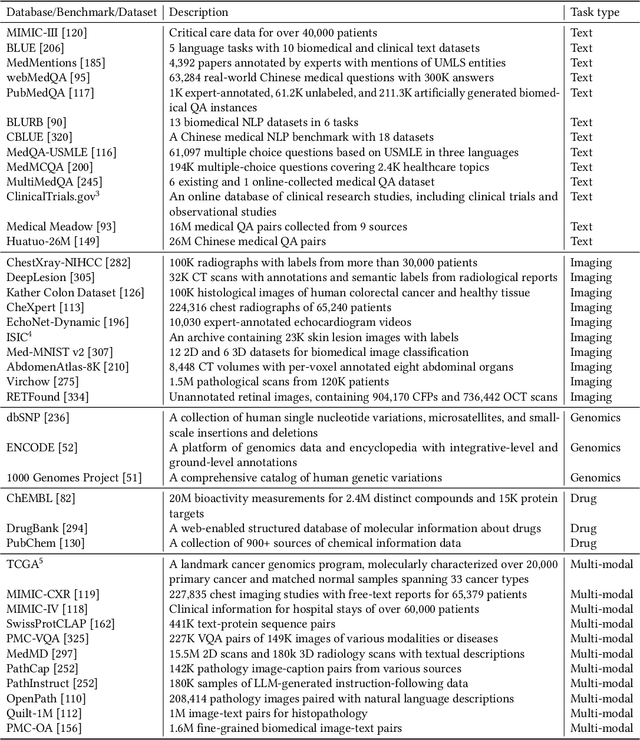
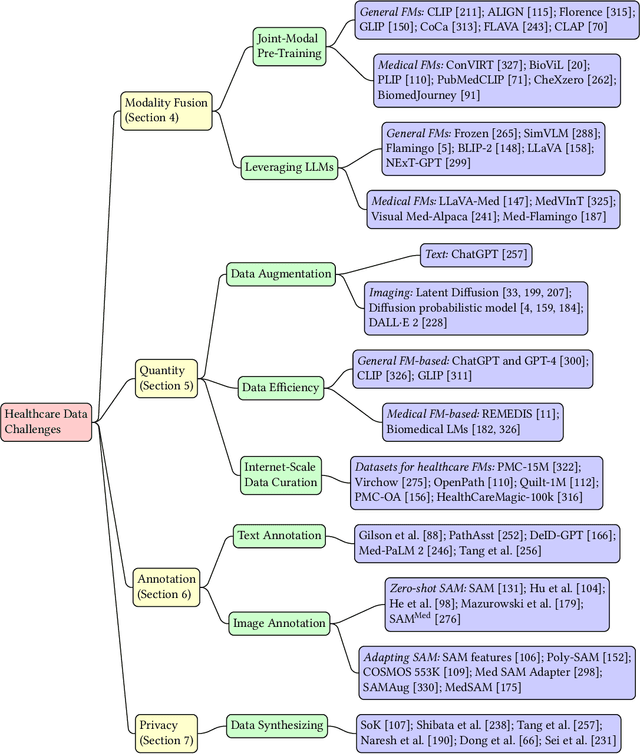
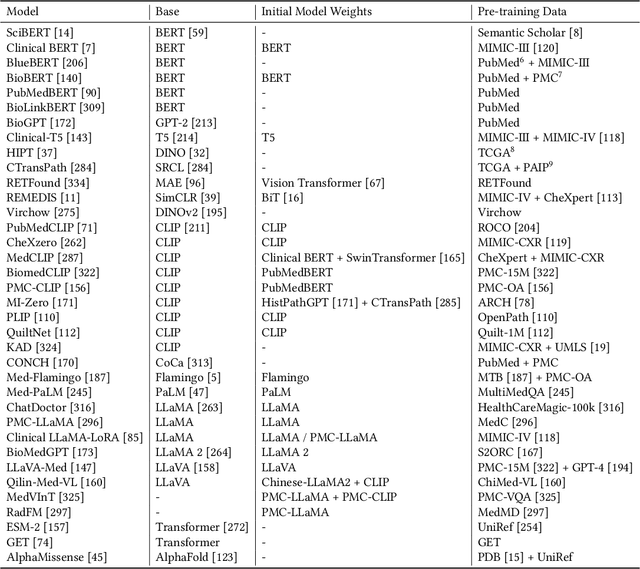
Abstract:The advent of foundation models (FMs) as an emerging suite of AI techniques has struck a wave of opportunities in computational healthcare. The interactive nature of these models, guided by pre-training data and human instructions, has ignited a data-centric AI paradigm that emphasizes better data characterization, quality, and scale. In healthcare AI, obtaining and processing high-quality clinical data records has been a longstanding challenge, ranging from data quantity, annotation, patient privacy, and ethics. In this survey, we investigate a wide range of data-centric approaches in the FM era (from model pre-training to inference) towards improving the healthcare workflow. We discuss key perspectives in AI security, assessment, and alignment with human values. Finally, we offer a promising outlook of FM-based analytics to enhance the performance of patient outcome and clinical workflow in the evolving landscape of healthcare and medicine. We provide an up-to-date list of healthcare-related foundation models and datasets at https://github.com/Yunkun-Zhang/Data-Centric-FM-Healthcare .
Acceleration Estimation of Signal Propagation Path Length Changes for Wireless Sensing
Dec 30, 2023



Abstract:As indoor applications grow in diversity, wireless sensing, vital in areas like localization and activity recognition, is attracting renewed interest. Indoor wireless sensing relies on signal processing, particularly channel state information (CSI) based signal parameter estimation. Nonetheless, regarding reflected signals induced by dynamic human targets, no satisfactory algorithm yet exists for estimating the acceleration of dynamic path length change (DPLC), which is crucial for various sensing tasks in this context. Hence, this paper proposes DP-AcE, a CSI-based DPLC acceleration estimation algorithm. We first model the relationship between the phase difference of adjacent CSI measurements and the DPLC's acceleration. Unlike existing works assuming constant velocity, DP-AcE considers both velocity and acceleration, yielding a more accurate and objective representation. Using this relationship, an algorithm combining scaling with Fourier transform is proposed to realize acceleration estimation. We evaluate DP-AcE via the acceleration estimation and acceleration-based fall detection with the collected CSI. Experimental results reveal that, using distance as the metric, DP-AcE achieves a median acceleration estimation percentage error of 4.38%. Furthermore, in multi-target scenarios, the fall detection achieves an average true positive rate of 89.56% and a false positive rate of 11.78%, demonstrating its importance in enhancing indoor wireless sensing capabilities.
SegRap2023: A Benchmark of Organs-at-Risk and Gross Tumor Volume Segmentation for Radiotherapy Planning of Nasopharyngeal Carcinoma
Dec 15, 2023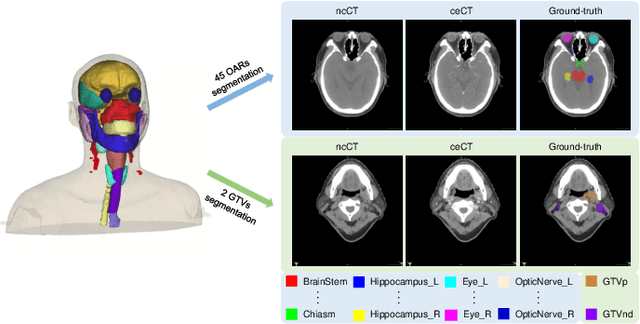
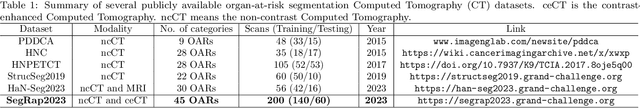
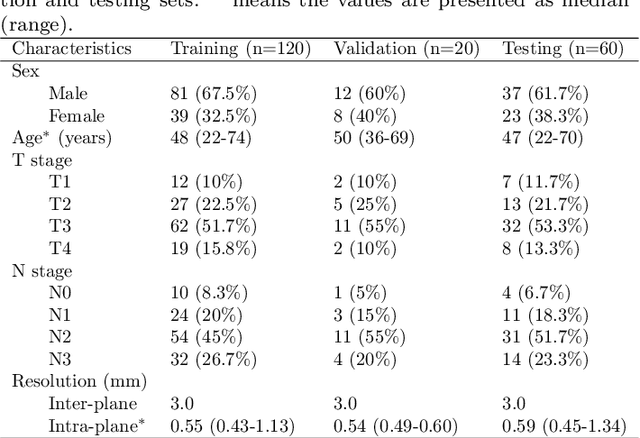
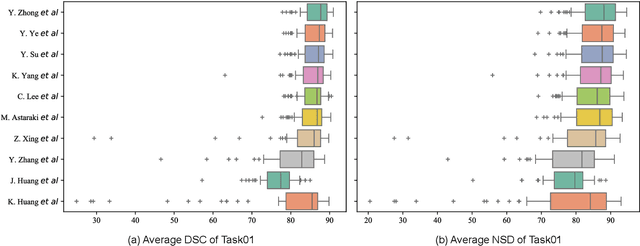
Abstract:Radiation therapy is a primary and effective NasoPharyngeal Carcinoma (NPC) treatment strategy. The precise delineation of Gross Tumor Volumes (GTVs) and Organs-At-Risk (OARs) is crucial in radiation treatment, directly impacting patient prognosis. Previously, the delineation of GTVs and OARs was performed by experienced radiation oncologists. Recently, deep learning has achieved promising results in many medical image segmentation tasks. However, for NPC OARs and GTVs segmentation, few public datasets are available for model development and evaluation. To alleviate this problem, the SegRap2023 challenge was organized in conjunction with MICCAI2023 and presented a large-scale benchmark for OAR and GTV segmentation with 400 Computed Tomography (CT) scans from 200 NPC patients, each with a pair of pre-aligned non-contrast and contrast-enhanced CT scans. The challenge's goal was to segment 45 OARs and 2 GTVs from the paired CT scans. In this paper, we detail the challenge and analyze the solutions of all participants. The average Dice similarity coefficient scores for all submissions ranged from 76.68\% to 86.70\%, and 70.42\% to 73.44\% for OARs and GTVs, respectively. We conclude that the segmentation of large-size OARs is well-addressed, and more efforts are needed for GTVs and small-size or thin-structure OARs. The benchmark will remain publicly available here: https://segrap2023.grand-challenge.org
Text-guided Foundation Model Adaptation for Pathological Image Classification
Jul 27, 2023



Abstract:The recent surge of foundation models in computer vision and natural language processing opens up perspectives in utilizing multi-modal clinical data to train large models with strong generalizability. Yet pathological image datasets often lack biomedical text annotation and enrichment. Guiding data-efficient image diagnosis from the use of biomedical text knowledge becomes a substantial interest. In this paper, we propose to Connect Image and Text Embeddings (CITE) to enhance pathological image classification. CITE injects text insights gained from language models pre-trained with a broad range of biomedical texts, leading to adapt foundation models towards pathological image understanding. Through extensive experiments on the PatchGastric stomach tumor pathological image dataset, we demonstrate that CITE achieves leading performance compared with various baselines especially when training data is scarce. CITE offers insights into leveraging in-domain text knowledge to reinforce data-efficient pathological image classification. Code is available at https://github.com/Yunkun-Zhang/CITE.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge