Lilong Wang
Beyond Static Tools: Test-Time Tool Evolution for Scientific Reasoning
Jan 12, 2026Abstract:The central challenge of AI for Science is not reasoning alone, but the ability to create computational methods in an open-ended scientific world. Existing LLM-based agents rely on static, pre-defined tool libraries, a paradigm that fundamentally fails in scientific domains where tools are sparse, heterogeneous, and intrinsically incomplete. In this paper, we propose Test-Time Tool Evolution (TTE), a new paradigm that enables agents to synthesize, verify, and evolve executable tools during inference. By transforming tools from fixed resources into problem-driven artifacts, TTE overcomes the rigidity and long-tail limitations of static tool libraries. To facilitate rigorous evaluation, we introduce SciEvo, a benchmark comprising 1,590 scientific reasoning tasks supported by 925 automatically evolved tools. Extensive experiments show that TTE achieves state-of-the-art performance in both accuracy and tool efficiency, while enabling effective cross-domain adaptation of computational tools. The code and benchmark have been released at https://github.com/lujiaxuan0520/Test-Time-Tool-Evol.
SCP: Accelerating Discovery with a Global Web of Autonomous Scientific Agents
Dec 30, 2025Abstract:We introduce SCP: the Science Context Protocol, an open-source standard designed to accelerate discovery by enabling a global network of autonomous scientific agents. SCP is built on two foundational pillars: (1) Unified Resource Integration: At its core, SCP provides a universal specification for describing and invoking scientific resources, spanning software tools, models, datasets, and physical instruments. This protocol-level standardization enables AI agents and applications to discover, call, and compose capabilities seamlessly across disparate platforms and institutional boundaries. (2) Orchestrated Experiment Lifecycle Management: SCP complements the protocol with a secure service architecture, which comprises a centralized SCP Hub and federated SCP Servers. This architecture manages the complete experiment lifecycle (registration, planning, execution, monitoring, and archival), enforces fine-grained authentication and authorization, and orchestrates traceable, end-to-end workflows that bridge computational and physical laboratories. Based on SCP, we have constructed a scientific discovery platform that offers researchers and agents a large-scale ecosystem of more than 1,600 tool resources. Across diverse use cases, SCP facilitates secure, large-scale collaboration between heterogeneous AI systems and human researchers while significantly reducing integration overhead and enhancing reproducibility. By standardizing scientific context and tool orchestration at the protocol level, SCP establishes essential infrastructure for scalable, multi-institution, agent-driven science.
BrainMVP: Multi-modal Vision Pre-training for Brain Image Analysis using Multi-parametric MRI
Oct 14, 2024



Abstract:Accurate diagnosis of brain abnormalities is greatly enhanced by the inclusion of complementary multi-parametric MRI imaging data. There is significant potential to develop a universal pre-training model that can be quickly adapted for image modalities and various clinical scenarios. However, current models often rely on uni-modal image data, neglecting the cross-modal correlations among different image modalities or struggling to scale up pre-training in the presence of missing modality data. In this paper, we propose BrainMVP, a multi-modal vision pre-training framework for brain image analysis using multi-parametric MRI scans. First, we collect 16,022 brain MRI scans (over 2.4 million images), encompassing eight MRI modalities sourced from a diverse range of centers and devices. Then, a novel pre-training paradigm is proposed for the multi-modal MRI data, addressing the issue of missing modalities and achieving multi-modal information fusion. Cross-modal reconstruction is explored to learn distinctive brain image embeddings and efficient modality fusion capabilities. A modality-wise data distillation module is proposed to extract the essence representation of each MR image modality for both the pre-training and downstream application purposes. Furthermore, we introduce a modality-aware contrastive learning module to enhance the cross-modality association within a study. Extensive experiments on downstream tasks demonstrate superior performance compared to state-of-the-art pre-training methods in the medical domain, with Dice Score improvement of 0.28%-14.47% across six segmentation benchmarks and a consistent accuracy improvement of 0.65%-18.07% in four individual classification tasks.
Cost-effective Instruction Learning for Pathology Vision and Language Analysis
Jul 25, 2024
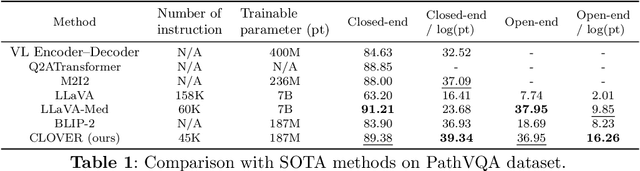
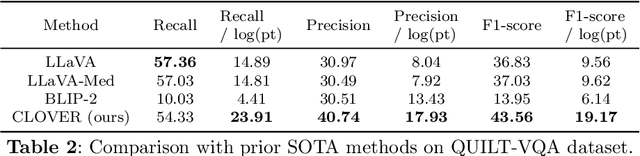

Abstract:The advent of vision-language models fosters the interactive conversations between AI-enabled models and humans. Yet applying these models into clinics must deal with daunting challenges around large-scale training data, financial, and computational resources. Here we propose a cost-effective instruction learning framework for conversational pathology named as CLOVER. CLOVER only trains a lightweight module and uses instruction tuning while freezing the parameters of the large language model. Instead of using costly GPT-4, we propose well-designed prompts on GPT-3.5 for building generation-based instructions, emphasizing the utility of pathological knowledge derived from the Internet source. To augment the use of instructions, we construct a high-quality set of template-based instructions in the context of digital pathology. From two benchmark datasets, our findings reveal the strength of hybrid-form instructions in the visual question-answer in pathology. Extensive results show the cost-effectiveness of CLOVER in answering both open-ended and closed-ended questions, where CLOVER outperforms strong baselines that possess 37 times more training parameters and use instruction data generated from GPT-4. Through the instruction tuning, CLOVER exhibits robustness of few-shot learning in the external clinical dataset. These findings demonstrate that cost-effective modeling of CLOVER could accelerate the adoption of rapid conversational applications in the landscape of digital pathology.
MedFMC: A Real-world Dataset and Benchmark For Foundation Model Adaptation in Medical Image Classification
Jun 16, 2023Abstract:Foundation models, often pre-trained with large-scale data, have achieved paramount success in jump-starting various vision and language applications. Recent advances further enable adapting foundation models in downstream tasks efficiently using only a few training samples, e.g., in-context learning. Yet, the application of such learning paradigms in medical image analysis remains scarce due to the shortage of publicly accessible data and benchmarks. In this paper, we aim at approaches adapting the foundation models for medical image classification and present a novel dataset and benchmark for the evaluation, i.e., examining the overall performance of accommodating the large-scale foundation models downstream on a set of diverse real-world clinical tasks. We collect five sets of medical imaging data from multiple institutes targeting a variety of real-world clinical tasks (22,349 images in total), i.e., thoracic diseases screening in X-rays, pathological lesion tissue screening, lesion detection in endoscopy images, neonatal jaundice evaluation, and diabetic retinopathy grading. Results of multiple baseline methods are demonstrated using the proposed dataset from both accuracy and cost-effective perspectives.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge