Xiaoqiang Liu
3D-Aware Implicit Motion Control for View-Adaptive Human Video Generation
Feb 03, 2026Abstract:Existing methods for human motion control in video generation typically rely on either 2D poses or explicit 3D parametric models (e.g., SMPL) as control signals. However, 2D poses rigidly bind motion to the driving viewpoint, precluding novel-view synthesis. Explicit 3D models, though structurally informative, suffer from inherent inaccuracies (e.g., depth ambiguity and inaccurate dynamics) which, when used as a strong constraint, override the powerful intrinsic 3D awareness of large-scale video generators. In this work, we revisit motion control from a 3D-aware perspective, advocating for an implicit, view-agnostic motion representation that naturally aligns with the generator's spatial priors rather than depending on externally reconstructed constraints. We introduce 3DiMo, which jointly trains a motion encoder with a pretrained video generator to distill driving frames into compact, view-agnostic motion tokens, injected semantically via cross-attention. To foster 3D awareness, we train with view-rich supervision (i.e., single-view, multi-view, and moving-camera videos), forcing motion consistency across diverse viewpoints. Additionally, we use auxiliary geometric supervision that leverages SMPL only for early initialization and is annealed to zero, enabling the model to transition from external 3D guidance to learning genuine 3D spatial motion understanding from the data and the generator's priors. Experiments confirm that 3DiMo faithfully reproduces driving motions with flexible, text-driven camera control, significantly surpassing existing methods in both motion fidelity and visual quality.
From Inpainting to Editing: A Self-Bootstrapping Framework for Context-Rich Visual Dubbing
Dec 31, 2025Abstract:Audio-driven visual dubbing aims to synchronize a video's lip movements with new speech, but is fundamentally challenged by the lack of ideal training data: paired videos where only a subject's lip movements differ while all other visual conditions are identical. Existing methods circumvent this with a mask-based inpainting paradigm, where an incomplete visual conditioning forces models to simultaneously hallucinate missing content and sync lips, leading to visual artifacts, identity drift, and poor synchronization. In this work, we propose a novel self-bootstrapping framework that reframes visual dubbing from an ill-posed inpainting task into a well-conditioned video-to-video editing problem. Our approach employs a Diffusion Transformer, first as a data generator, to synthesize ideal training data: a lip-altered companion video for each real sample, forming visually aligned video pairs. A DiT-based audio-driven editor is then trained on these pairs end-to-end, leveraging the complete and aligned input video frames to focus solely on precise, audio-driven lip modifications. This complete, frame-aligned input conditioning forms a rich visual context for the editor, providing it with complete identity cues, scene interactions, and continuous spatiotemporal dynamics. Leveraging this rich context fundamentally enables our method to achieve highly accurate lip sync, faithful identity preservation, and exceptional robustness against challenging in-the-wild scenarios. We further introduce a timestep-adaptive multi-phase learning strategy as a necessary component to disentangle conflicting editing objectives across diffusion timesteps, thereby facilitating stable training and yielding enhanced lip synchronization and visual fidelity. Additionally, we propose ContextDubBench, a comprehensive benchmark dataset for robust evaluation in diverse and challenging practical application scenarios.
KlingAvatar 2.0 Technical Report
Dec 15, 2025Abstract:Avatar video generation models have achieved remarkable progress in recent years. However, prior work exhibits limited efficiency in generating long-duration high-resolution videos, suffering from temporal drifting, quality degradation, and weak prompt following as video length increases. To address these challenges, we propose KlingAvatar 2.0, a spatio-temporal cascade framework that performs upscaling in both spatial resolution and temporal dimension. The framework first generates low-resolution blueprint video keyframes that capture global semantics and motion, and then refines them into high-resolution, temporally coherent sub-clips using a first-last frame strategy, while retaining smooth temporal transitions in long-form videos. To enhance cross-modal instruction fusion and alignment in extended videos, we introduce a Co-Reasoning Director composed of three modality-specific large language model (LLM) experts. These experts reason about modality priorities and infer underlying user intent, converting inputs into detailed storylines through multi-turn dialogue. A Negative Director further refines negative prompts to improve instruction alignment. Building on these components, we extend the framework to support ID-specific multi-character control. Extensive experiments demonstrate that our model effectively addresses the challenges of efficient, multimodally aligned long-form high-resolution video generation, delivering enhanced visual clarity, realistic lip-teeth rendering with accurate lip synchronization, strong identity preservation, and coherent multimodal instruction following.
Kling-Avatar: Grounding Multimodal Instructions for Cascaded Long-Duration Avatar Animation Synthesis
Sep 11, 2025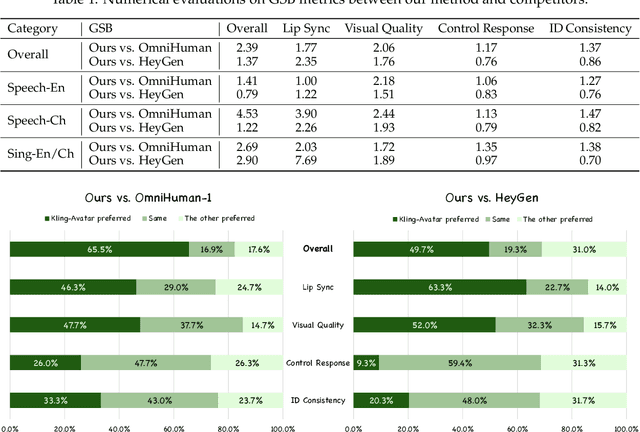
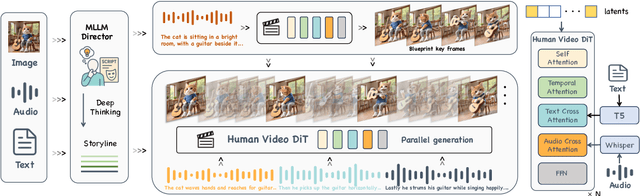


Abstract:Recent advances in audio-driven avatar video generation have significantly enhanced audio-visual realism. However, existing methods treat instruction conditioning merely as low-level tracking driven by acoustic or visual cues, without modeling the communicative purpose conveyed by the instructions. This limitation compromises their narrative coherence and character expressiveness. To bridge this gap, we introduce Kling-Avatar, a novel cascaded framework that unifies multimodal instruction understanding with photorealistic portrait generation. Our approach adopts a two-stage pipeline. In the first stage, we design a multimodal large language model (MLLM) director that produces a blueprint video conditioned on diverse instruction signals, thereby governing high-level semantics such as character motion and emotions. In the second stage, guided by blueprint keyframes, we generate multiple sub-clips in parallel using a first-last frame strategy. This global-to-local framework preserves fine-grained details while faithfully encoding the high-level intent behind multimodal instructions. Our parallel architecture also enables fast and stable generation of long-duration videos, making it suitable for real-world applications such as digital human livestreaming and vlogging. To comprehensively evaluate our method, we construct a benchmark of 375 curated samples covering diverse instructions and challenging scenarios. Extensive experiments demonstrate that Kling-Avatar is capable of generating vivid, fluent, long-duration videos at up to 1080p and 48 fps, achieving superior performance in lip synchronization accuracy, emotion and dynamic expressiveness, instruction controllability, identity preservation, and cross-domain generalization. These results establish Kling-Avatar as a new benchmark for semantically grounded, high-fidelity audio-driven avatar synthesis.
MIDAS: Multimodal Interactive Digital-humAn Synthesis via Real-time Autoregressive Video Generation
Aug 28, 2025Abstract:Recently, interactive digital human video generation has attracted widespread attention and achieved remarkable progress. However, building such a practical system that can interact with diverse input signals in real time remains challenging to existing methods, which often struggle with heavy computational cost and limited controllability. In this work, we introduce an autoregressive video generation framework that enables interactive multimodal control and low-latency extrapolation in a streaming manner. With minimal modifications to a standard large language model (LLM), our framework accepts multimodal condition encodings including audio, pose, and text, and outputs spatially and semantically coherent representations to guide the denoising process of a diffusion head. To support this, we construct a large-scale dialogue dataset of approximately 20,000 hours from multiple sources, providing rich conversational scenarios for training. We further introduce a deep compression autoencoder with up to 64$\times$ reduction ratio, which effectively alleviates the long-horizon inference burden of the autoregressive model. Extensive experiments on duplex conversation, multilingual human synthesis, and interactive world model highlight the advantages of our approach in low latency, high efficiency, and fine-grained multimodal controllability.
GGTalker: Talking Head Systhesis with Generalizable Gaussian Priors and Identity-Specific Adaptation
Jun 26, 2025Abstract:Creating high-quality, generalizable speech-driven 3D talking heads remains a persistent challenge. Previous methods achieve satisfactory results for fixed viewpoints and small-scale audio variations, but they struggle with large head rotations and out-of-distribution (OOD) audio. Moreover, they are constrained by the need for time-consuming, identity-specific training. We believe the core issue lies in the lack of sufficient 3D priors, which limits the extrapolation capabilities of synthesized talking heads. To address this, we propose GGTalker, which synthesizes talking heads through a combination of generalizable priors and identity-specific adaptation. We introduce a two-stage Prior-Adaptation training strategy to learn Gaussian head priors and adapt to individual characteristics. We train Audio-Expression and Expression-Visual priors to capture the universal patterns of lip movements and the general distribution of head textures. During the Customized Adaptation, individual speaking styles and texture details are precisely modeled. Additionally, we introduce a color MLP to generate fine-grained, motion-aligned textures and a Body Inpainter to blend rendered results with the background, producing indistinguishable, photorealistic video frames. Comprehensive experiments show that GGTalker achieves state-of-the-art performance in rendering quality, 3D consistency, lip-sync accuracy, and training efficiency.
OmniSync: Towards Universal Lip Synchronization via Diffusion Transformers
May 27, 2025Abstract:Lip synchronization is the task of aligning a speaker's lip movements in video with corresponding speech audio, and it is essential for creating realistic, expressive video content. However, existing methods often rely on reference frames and masked-frame inpainting, which limit their robustness to identity consistency, pose variations, facial occlusions, and stylized content. In addition, since audio signals provide weaker conditioning than visual cues, lip shape leakage from the original video will affect lip sync quality. In this paper, we present OmniSync, a universal lip synchronization framework for diverse visual scenarios. Our approach introduces a mask-free training paradigm using Diffusion Transformer models for direct frame editing without explicit masks, enabling unlimited-duration inference while maintaining natural facial dynamics and preserving character identity. During inference, we propose a flow-matching-based progressive noise initialization to ensure pose and identity consistency, while allowing precise mouth-region editing. To address the weak conditioning signal of audio, we develop a Dynamic Spatiotemporal Classifier-Free Guidance (DS-CFG) mechanism that adaptively adjusts guidance strength over time and space. We also establish the AIGC-LipSync Benchmark, the first evaluation suite for lip synchronization in diverse AI-generated videos. Extensive experiments demonstrate that OmniSync significantly outperforms prior methods in both visual quality and lip sync accuracy, achieving superior results in both real-world and AI-generated videos.
MambaCapsule: Towards Transparent Cardiac Disease Diagnosis with Electrocardiography Using Mamba Capsule Network
Jul 30, 2024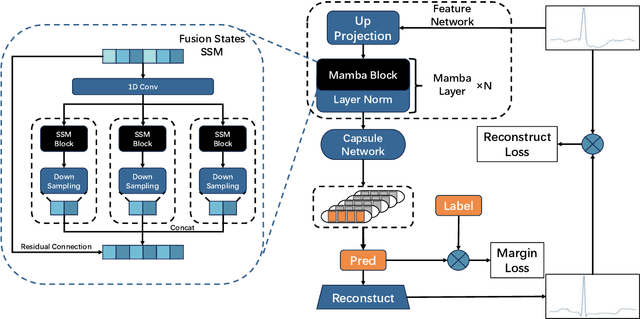
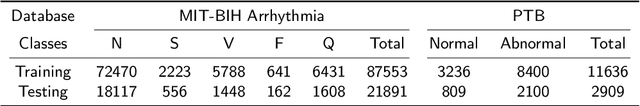
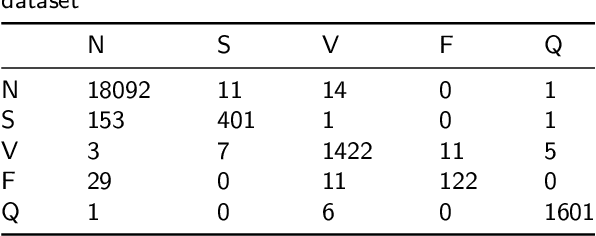
Abstract:Cardiac arrhythmia, a condition characterized by irregular heartbeats, often serves as an early indication of various heart ailments. With the advent of deep learning, numerous innovative models have been introduced for diagnosing arrhythmias using Electrocardiogram (ECG) signals. However, recent studies solely focus on the performance of models, neglecting the interpretation of their results. This leads to a considerable lack of transparency, posing a significant risk in the actual diagnostic process. To solve this problem, this paper introduces MambaCapsule, a deep neural networks for ECG arrhythmias classification, which increases the explainability of the model while enhancing the accuracy.Our model utilizes Mamba for feature extraction and Capsule networks for prediction, providing not only a confidence score but also signal features. Akin to the processing mechanism of human brain, the model learns signal features and their relationship between them by reconstructing ECG signals in the predicted selection. The model evaluation was conducted on MIT-BIH and PTB dataset, following the AAMI standard. MambaCapsule has achieved a total accuracy of 99.54% and 99.59% on the test sets respectively. These results demonstrate the promising performance of under the standard test protocol.
LivePortrait: Efficient Portrait Animation with Stitching and Retargeting Control
Jul 03, 2024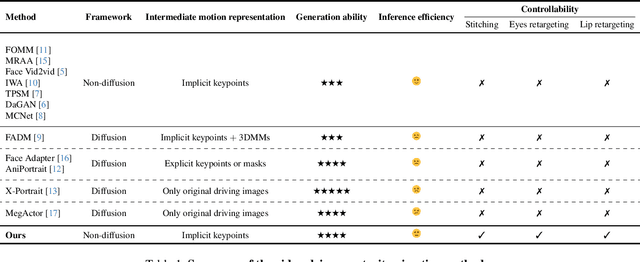
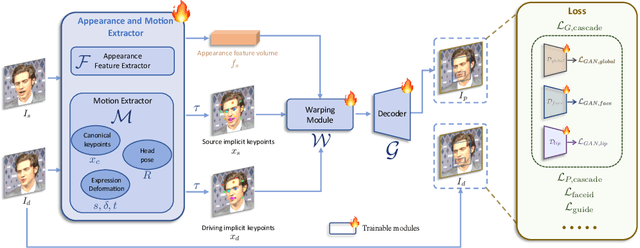
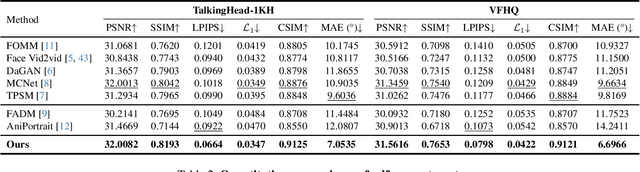
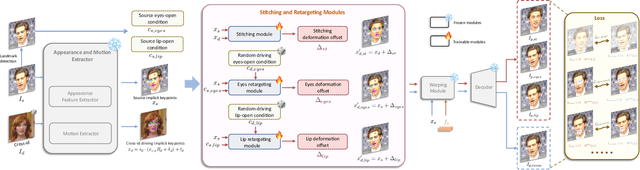
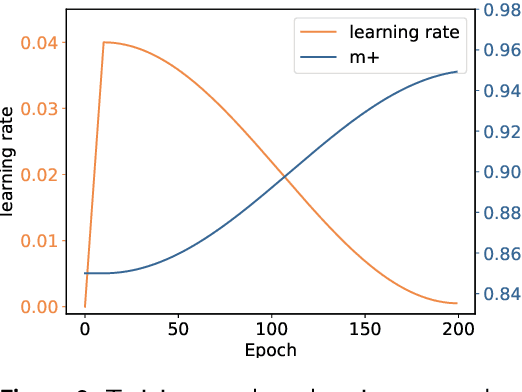
Abstract:Portrait Animation aims to synthesize a lifelike video from a single source image, using it as an appearance reference, with motion (i.e., facial expressions and head pose) derived from a driving video, audio, text, or generation. Instead of following mainstream diffusion-based methods, we explore and extend the potential of the implicit-keypoint-based framework, which effectively balances computational efficiency and controllability. Building upon this, we develop a video-driven portrait animation framework named LivePortrait with a focus on better generalization, controllability, and efficiency for practical usage. To enhance the generation quality and generalization ability, we scale up the training data to about 69 million high-quality frames, adopt a mixed image-video training strategy, upgrade the network architecture, and design better motion transformation and optimization objectives. Additionally, we discover that compact implicit keypoints can effectively represent a kind of blendshapes and meticulously propose a stitching and two retargeting modules, which utilize a small MLP with negligible computational overhead, to enhance the controllability. Experimental results demonstrate the efficacy of our framework even compared to diffusion-based methods. The generation speed remarkably reaches 12.8ms on an RTX 4090 GPU with PyTorch. The inference code and models are available at https://github.com/KwaiVGI/LivePortrait
The Application of ChatGPT in Responding to Questions Related to the Boston Bowel Preparation Scale
Feb 13, 2024



Abstract:Background: Colonoscopy, a crucial diagnostic tool in gastroenterology, depends heavily on superior bowel preparation. ChatGPT, a large language model with emergent intelligence which also exhibits potential in medical applications. This study aims to assess the accuracy and consistency of ChatGPT in using the Boston Bowel Preparation Scale (BBPS) for colonoscopy assessment. Methods: We retrospectively collected 233 colonoscopy images from 2020 to 2023. These images were evaluated using the BBPS by 3 senior endoscopists and 3 novice endoscopists. Additionally, ChatGPT also assessed these images, having been divided into three groups and undergone specific Fine-tuning. Consistency was evaluated through two rounds of testing. Results: In the initial round, ChatGPT's accuracy varied between 48.93% and 62.66%, trailing the endoscopists' accuracy of 76.68% to 77.83%. Kappa values for ChatGPT was between 0.52 and 0.53, compared to 0.75 to 0.87 for the endoscopists. Conclusion: While ChatGPT shows promise in bowel preparation scoring, it currently does not match the accuracy and consistency of experienced endoscopists. Future research should focus on in-depth Fine-tuning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge