Mengzhang Li
MedFMC: A Real-world Dataset and Benchmark For Foundation Model Adaptation in Medical Image Classification
Jun 16, 2023Abstract:Foundation models, often pre-trained with large-scale data, have achieved paramount success in jump-starting various vision and language applications. Recent advances further enable adapting foundation models in downstream tasks efficiently using only a few training samples, e.g., in-context learning. Yet, the application of such learning paradigms in medical image analysis remains scarce due to the shortage of publicly accessible data and benchmarks. In this paper, we aim at approaches adapting the foundation models for medical image classification and present a novel dataset and benchmark for the evaluation, i.e., examining the overall performance of accommodating the large-scale foundation models downstream on a set of diverse real-world clinical tasks. We collect five sets of medical imaging data from multiple institutes targeting a variety of real-world clinical tasks (22,349 images in total), i.e., thoracic diseases screening in X-rays, pathological lesion tissue screening, lesion detection in endoscopy images, neonatal jaundice evaluation, and diabetic retinopathy grading. Results of multiple baseline methods are demonstrated using the proposed dataset from both accuracy and cost-effective perspectives.
Automatic Data Augmentation for 3D Medical Image Segmentation
Oct 07, 2020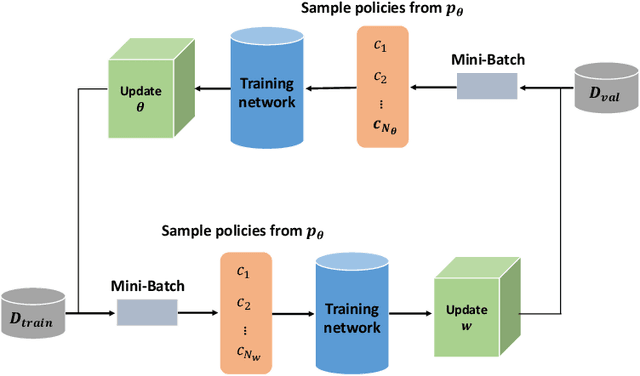
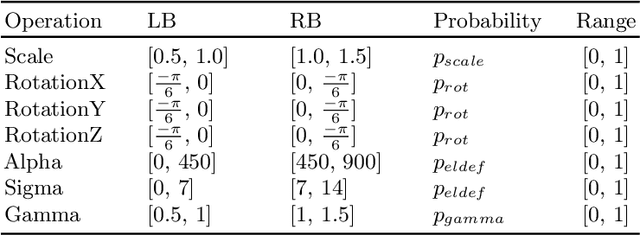
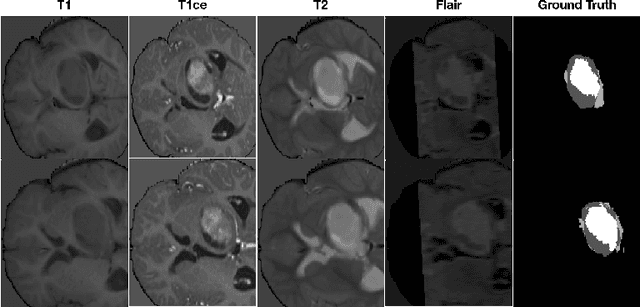
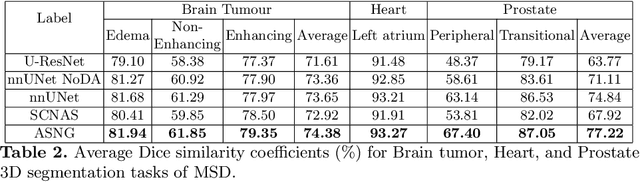
Abstract:Data augmentation is an effective and universal technique for improving generalization performance of deep neural networks. It could enrich diversity of training samples that is essential in medical image segmentation tasks because 1) the scale of medical image dataset is typically smaller, which may increase the risk of overfitting; 2) the shape and modality of different objects such as organs or tumors are unique, thus requiring customized data augmentation policy. However, most data augmentation implementations are hand-crafted and suboptimal in medical image processing. To fully exploit the potential of data augmentation, we propose an efficient algorithm to automatically search for the optimal augmentation strategies. We formulate the coupled optimization w.r.t. network weights and augmentation parameters into a differentiable form by means of stochastic relaxation. This formulation allows us to apply alternative gradient-based methods to solve it, i.e. stochastic natural gradient method with adaptive step-size. To the best of our knowledge, it is the first time that differentiable automatic data augmentation is employed in medical image segmentation tasks. Our numerical experiments demonstrate that the proposed approach significantly outperforms existing build-in data augmentation of state-of-the-art models.
3D Graph Convolutional Networks with Temporal Graphs: A Spatial Information Free Framework For Traffic Forecasting
Mar 03, 2019

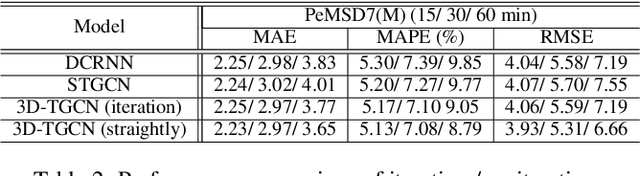
Abstract:Spatio-temporal prediction plays an important role in many application areas especially in traffic domain. However, due to complicated spatio-temporal dependency and high non-linear dynamics in road networks, traffic prediction task is still challenging. Existing works either exhibit heavy training cost or fail to accurately capture the spatio-temporal patterns, also ignore the correlation between distant roads that share the similar patterns. In this paper, we propose a novel deep learning framework to overcome these issues: 3D Temporal Graph Convolutional Networks (3D-TGCN). Two novel components of our model are introduced. (1) Instead of constructing the road graph based on spatial information, we learn it by comparing the similarity between time series for each road, thus providing a spatial information free framework. (2) We propose an original 3D graph convolution model to model the spatio-temporal data more accurately. Empirical results show that 3D-TGCN could outperform state-of-the-art baselines.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge