Dequan Wang
daVinci-Agency: Unlocking Long-Horizon Agency Data-Efficiently
Feb 02, 2026Abstract:While Large Language Models (LLMs) excel at short-term tasks, scaling them to long-horizon agentic workflows remains challenging. The core bottleneck lies in the scarcity of training data that captures authentic long-dependency structures and cross-stage evolutionary dynamics--existing synthesis methods either confine to single-feature scenarios constrained by model distribution, or incur prohibitive human annotation costs, failing to provide scalable, high-quality supervision. We address this by reconceptualizing data synthesis through the lens of real-world software evolution. Our key insight: Pull Request (PR) sequences naturally embody the supervision signals for long-horizon learning. They decompose complex objectives into verifiable submission units, maintain functional coherence across iterations, and encode authentic refinement patterns through bug-fix histories. Building on this, we propose daVinci-Agency, which systematically mines structured supervision from chain-of-PRs through three interlocking mechanisms: (1) progressive task decomposition via continuous commits, (2) long-term consistency enforcement through unified functional objectives, and (3) verifiable refinement from authentic bug-fix trajectories. Unlike synthetic trajectories that treat each step independently, daVinci-Agency's PR-grounded structure inherently preserves the causal dependencies and iterative refinements essential for teaching persistent goal-directed behavior and enables natural alignment with project-level, full-cycle task modeling. The resulting trajectories are substantial--averaging 85k tokens and 116 tool calls--yet remarkably data-efficient: fine-tuning GLM-4.6 on 239 daVinci-Agency samples yields broad improvements across benchmarks, notably achieving a 47% relative gain on Toolathlon. Beyond benchmark performance, our analysis confirms...
AgencyBench: Benchmarking the Frontiers of Autonomous Agents in 1M-Token Real-World Contexts
Jan 16, 2026Abstract:Large Language Models (LLMs) based autonomous agents demonstrate multifaceted capabilities to contribute substantially to economic production. However, existing benchmarks remain focused on single agentic capability, failing to capture long-horizon real-world scenarios. Moreover, the reliance on human-in-the-loop feedback for realistic tasks creates a scalability bottleneck, hindering automated rollout collection and evaluation. To bridge this gap, we introduce AgencyBench, a comprehensive benchmark derived from daily AI usage, evaluating 6 core agentic capabilities across 32 real-world scenarios, comprising 138 tasks with specific queries, deliverables, and rubrics. These scenarios require an average of 90 tool calls, 1 million tokens, and hours of execution time to resolve. To enable automated evaluation, we employ a user simulation agent to provide iterative feedback, and a Docker sandbox to conduct visual and functional rubric-based assessment. Experiments reveal that closed-source models significantly outperform open-source models (48.4% vs 32.1%). Further analysis reveals significant disparities across models in resource efficiency, feedback-driven self-correction, and specific tool-use preferences. Finally, we investigate the impact of agentic scaffolds, observing that proprietary models demonstrate superior performance within their native ecosystems (e.g., Claude-4.5-Opus via Claude-Agent-SDK), while open-source models exhibit distinct performance peaks, suggesting potential optimization for specific execution frameworks. AgencyBench serves as a critical testbed for next-generation agents, highlighting the necessity of co-optimizing model architecture with agentic frameworks. We believe this work sheds light on the future direction of autonomous agents, and we release the full benchmark and evaluation toolkit at https://github.com/GAIR-NLP/AgencyBench.
Saying the Unsaid: Revealing the Hidden Language of Multimodal Systems Through Telephone Games
Nov 12, 2025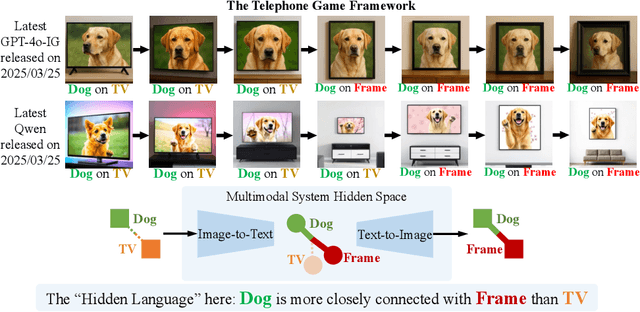
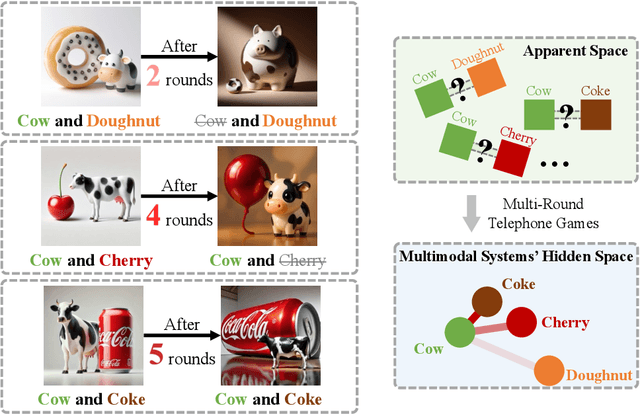
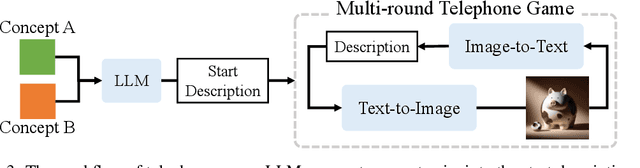

Abstract:Recent closed-source multimodal systems have made great advances, but their hidden language for understanding the world remains opaque because of their black-box architectures. In this paper, we use the systems' preference bias to study their hidden language: During the process of compressing the input images (typically containing multiple concepts) into texts and then reconstructing them into images, the systems' inherent preference bias introduces specific shifts in the outputs, disrupting the original input concept co-occurrence. We employ the multi-round "telephone game" to strategically leverage this bias. By observing the co-occurrence frequencies of concepts in telephone games, we quantitatively investigate the concept connection strength in the understanding of multimodal systems, i.e., "hidden language." We also contribute Telescope, a dataset of 10,000+ concept pairs, as the database of our telephone game framework. Our telephone game is test-time scalable: By iteratively running telephone games, we can construct a global map of concept connections in multimodal systems' understanding. Here we can identify preference bias inherited from training, assess generalization capability advancement, and discover more stable pathways for fragile concept connections. Furthermore, we use Reasoning-LLMs to uncover unexpected concept relationships that transcend textual and visual similarities, inferring how multimodal systems understand and simulate the world. This study offers a new perspective on the hidden language of multimodal systems and lays the foundation for future research on the interpretability and controllability of multimodal systems.
DatasetResearch: Benchmarking Agent Systems for Demand-Driven Dataset Discovery
Aug 09, 2025Abstract:The rapid advancement of large language models has fundamentally shifted the bottleneck in AI development from computational power to data availability-with countless valuable datasets remaining hidden across specialized repositories, research appendices, and domain platforms. As reasoning capabilities and deep research methodologies continue to evolve, a critical question emerges: can AI agents transcend conventional search to systematically discover any dataset that meets specific user requirements, enabling truly autonomous demand-driven data curation? We introduce DatasetResearch, the first comprehensive benchmark evaluating AI agents' ability to discover and synthesize datasets from 208 real-world demands across knowledge-intensive and reasoning-intensive tasks. Our tri-dimensional evaluation framework reveals a stark reality: even advanced deep research systems achieve only 22% score on our challenging DatasetResearch-pro subset, exposing the vast gap between current capabilities and perfect dataset discovery. Our analysis uncovers a fundamental dichotomy-search agents excel at knowledge tasks through retrieval breadth, while synthesis agents dominate reasoning challenges via structured generation-yet both catastrophically fail on "corner cases" outside existing distributions. These findings establish the first rigorous baseline for dataset discovery agents and illuminate the path toward AI systems capable of finding any dataset in the digital universe. Our benchmark and comprehensive analysis provide the foundation for the next generation of self-improving AI systems and are publicly available at https://github.com/GAIR-NLP/DatasetResearch.
MedForge: Building Medical Foundation Models Like Open Source Software Development
Feb 22, 2025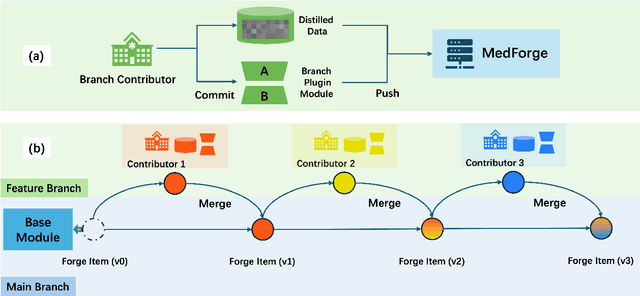
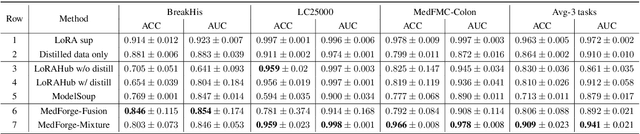
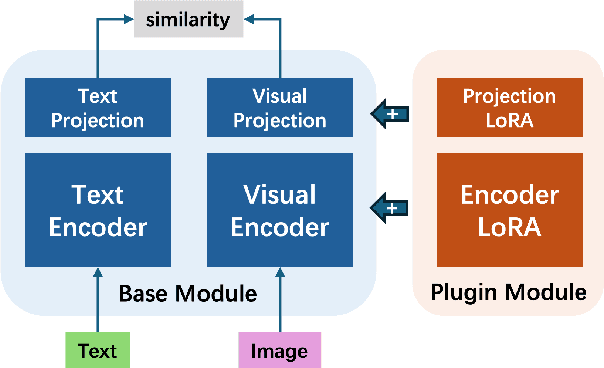
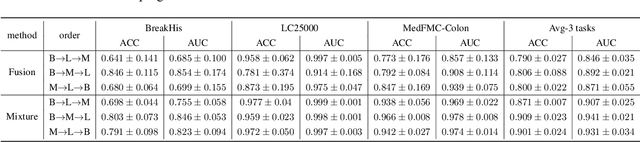
Abstract:Foundational models (FMs) have made significant strides in the healthcare domain. Yet the data silo challenge and privacy concern remain in healthcare systems, hindering safe medical data sharing and collaborative model development among institutions. The collection and curation of scalable clinical datasets increasingly become the bottleneck for training strong FMs. In this study, we propose Medical Foundation Models Merging (MedForge), a cooperative framework enabling a community-driven medical foundation model development, meanwhile preventing the information leakage of raw patient data and mitigating synchronization model development issues across clinical institutions. MedForge offers a bottom-up model construction mechanism by flexibly merging task-specific Low-Rank Adaptation (LoRA) modules, which can adapt to downstream tasks while retaining original model parameters. Through an asynchronous LoRA module integration scheme, the resulting composite model can progressively enhance its comprehensive performance on various clinical tasks. MedForge shows strong performance on multiple clinical datasets (e.g., breast cancer, lung cancer, and colon cancer) collected from different institutions. Our major findings highlight the value of collaborative foundation models in advancing multi-center clinical collaboration effectively and cohesively. Our code is publicly available at https://github.com/TanZheling/MedForge.
Lost in Translation: Latent Concept Misalignment in Text-to-Image Diffusion Models
Aug 05, 2024Abstract:Advancements in text-to-image diffusion models have broadened extensive downstream practical applications, but such models often encounter misalignment issues between text and image. Taking the generation of a combination of two disentangled concepts as an example, say given the prompt "a tea cup of iced coke", existing models usually generate a glass cup of iced coke because the iced coke usually co-occurs with the glass cup instead of the tea one during model training. The root of such misalignment is attributed to the confusion in the latent semantic space of text-to-image diffusion models, and hence we refer to the "a tea cup of iced coke" phenomenon as Latent Concept Misalignment (LC-Mis). We leverage large language models (LLMs) to thoroughly investigate the scope of LC-Mis, and develop an automated pipeline for aligning the latent semantics of diffusion models to text prompts. Empirical assessments confirm the effectiveness of our approach, substantially reducing LC-Mis errors and enhancing the robustness and versatility of text-to-image diffusion models. The code and dataset are here: https://github.com/RossoneriZhao/iced_coke.
Dissecting Dissonance: Benchmarking Large Multimodal Models Against Self-Contradictory Instructions
Aug 05, 2024



Abstract:Large multimodal models (LMMs) excel in adhering to human instructions. However, self-contradictory instructions may arise due to the increasing trend of multimodal interaction and context length, which is challenging for language beginners and vulnerable populations. We introduce the Self-Contradictory Instructions benchmark to evaluate the capability of LMMs in recognizing conflicting commands. It comprises 20,000 conflicts, evenly distributed between language and vision paradigms. It is constructed by a novel automatic dataset creation framework, which expedites the process and enables us to encompass a wide range of instruction forms. Our comprehensive evaluation reveals current LMMs consistently struggle to identify multimodal instruction discordance due to a lack of self-awareness. Hence, we propose the Cognitive Awakening Prompting to inject cognition from external, largely enhancing dissonance detection. The dataset and code are here: https://selfcontradiction.github.io/.
Direct Preference Knowledge Distillation for Large Language Models
Jun 28, 2024



Abstract:In the field of large language models (LLMs), Knowledge Distillation (KD) is a critical technique for transferring capabilities from teacher models to student models. However, existing KD methods face limitations and challenges in distillation of LLMs, including efficiency and insufficient measurement capabilities of traditional KL divergence. It is shown that LLMs can serve as an implicit reward function, which we define as a supplement to KL divergence. In this work, we propose Direct Preference Knowledge Distillation (DPKD) for LLMs. DPKD utilizes distribution divergence to represent the preference loss and implicit reward function. We re-formulate KD of LLMs into two stages: first optimizing and objective consisting of implicit reward and reverse KL divergence and then improving the preference probability of teacher outputs over student outputs. We conducted experiments and analysis on various datasets with LLM parameters ranging from 120M to 13B and demonstrate the broad applicability and effectiveness of our DPKD approach. Meanwhile, we prove the value and effectiveness of the introduced implicit reward and output preference in KD through experiments and theoretical analysis. The DPKD method outperforms the baseline method in both output response precision and exact match percentage. Code and data are available at https://aka.ms/dpkd.
mmPlace: Robust Place Recognition with Intermediate Frequency Signal of Low-cost Single-chip Millimeter Wave Radar
Mar 07, 2024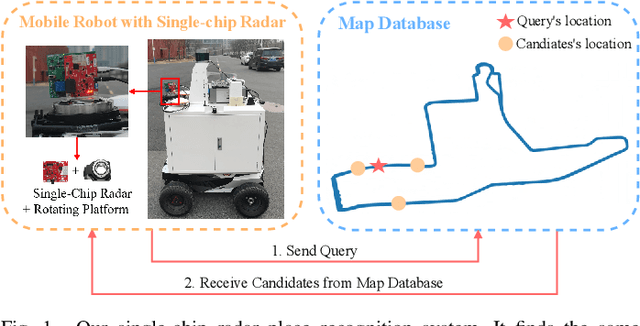

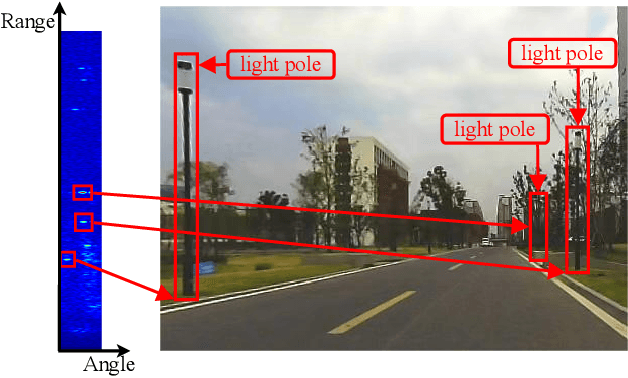
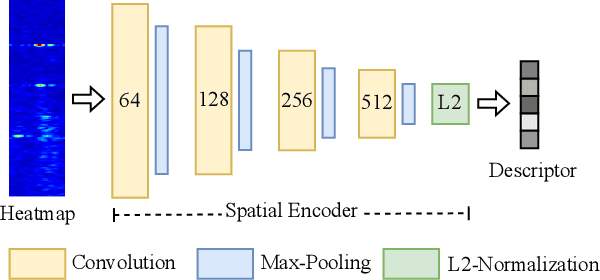
Abstract:Place recognition is crucial for tasks like loop-closure detection and re-localization. Single-chip millimeter wave radar (single-chip radar in short) emerges as a low-cost sensor option for place recognition, with the advantage of insensitivity to degraded visual environments. However, it encounters two challenges. Firstly, sparse point cloud from single-chip radar leads to poor performance when using current place recognition methods, which assume much denser data. Secondly, its performance significantly declines in scenarios involving rotational and lateral variations, due to limited overlap in its field of view (FOV). We propose mmPlace, a robust place recognition system to address these challenges. Specifically, mmPlace transforms intermediate frequency (IF) signal into range azimuth heatmap and employs a spatial encoder to extract features. Additionally, to improve the performance in scenarios involving rotational and lateral variations, mmPlace employs a rotating platform and concatenates heatmaps in a rotation cycle, effectively expanding the system's FOV. We evaluate mmPlace's performance on the milliSonic dataset, which is collected on the University of Science and Technology of China (USTC) campus, the city roads surrounding the campus, and an underground parking garage. The results demonstrate that mmPlace outperforms point cloud-based methods and achieves 87.37% recall@1 in scenarios involving rotational and lateral variations.
OpenMEDLab: An Open-source Platform for Multi-modality Foundation Models in Medicine
Mar 04, 2024



Abstract:The emerging trend of advancing generalist artificial intelligence, such as GPTv4 and Gemini, has reshaped the landscape of research (academia and industry) in machine learning and many other research areas. However, domain-specific applications of such foundation models (e.g., in medicine) remain untouched or often at their very early stages. It will require an individual set of transfer learning and model adaptation techniques by further expanding and injecting these models with domain knowledge and data. The development of such technologies could be largely accelerated if the bundle of data, algorithms, and pre-trained foundation models were gathered together and open-sourced in an organized manner. In this work, we present OpenMEDLab, an open-source platform for multi-modality foundation models. It encapsulates not only solutions of pioneering attempts in prompting and fine-tuning large language and vision models for frontline clinical and bioinformatic applications but also building domain-specific foundation models with large-scale multi-modal medical data. Importantly, it opens access to a group of pre-trained foundation models for various medical image modalities, clinical text, protein engineering, etc. Inspiring and competitive results are also demonstrated for each collected approach and model in a variety of benchmarks for downstream tasks. We welcome researchers in the field of medical artificial intelligence to continuously contribute cutting-edge methods and models to OpenMEDLab, which can be accessed via https://github.com/openmedlab.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge