Honghao Gao
Take Your Steps: Hierarchically Efficient Pulmonary Disease Screening via CT Volume Compression
Dec 03, 2024Abstract:Deep learning models are widely used to process Computed Tomography (CT) data in the automated screening of pulmonary diseases, significantly reducing the workload of physicians. However, the three-dimensional nature of CT volumes involves an excessive number of voxels, which significantly increases the complexity of model processing. Previous screening approaches often overlook this issue, which undoubtedly reduces screening efficiency. Towards efficient and effective screening, we design a hierarchical approach to reduce the computational cost of pulmonary disease screening. The new approach re-organizes the screening workflows into three steps. First, we propose a Computed Tomography Volume Compression (CTVC) method to select a small slice subset that comprehensively represents the whole CT volume. Second, the selected CT slices are used to detect pulmonary diseases coarsely via a lightweight classification model. Third, an uncertainty measurement strategy is applied to identify samples with low diagnostic confidence, which are re-detected by radiologists. Experiments on two public pulmonary disease datasets demonstrate that our approach achieves comparable accuracy and recall while reducing the time by 50%-70% compared with the counterparts using full CT volumes. Besides, we also found that our approach outperforms previous cutting-edge CTVC methods in retaining important indications after compression.
MambaCapsule: Towards Transparent Cardiac Disease Diagnosis with Electrocardiography Using Mamba Capsule Network
Jul 30, 2024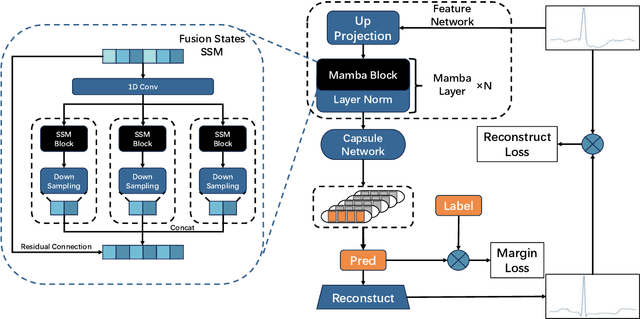
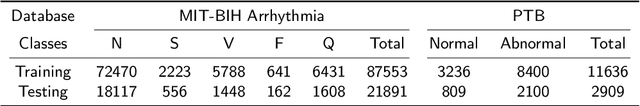
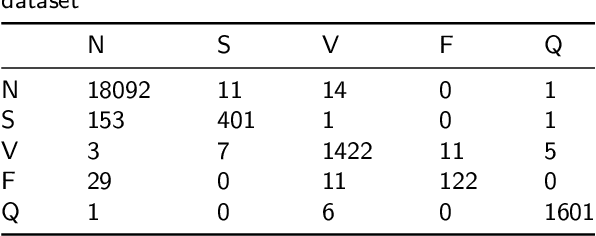
Abstract:Cardiac arrhythmia, a condition characterized by irregular heartbeats, often serves as an early indication of various heart ailments. With the advent of deep learning, numerous innovative models have been introduced for diagnosing arrhythmias using Electrocardiogram (ECG) signals. However, recent studies solely focus on the performance of models, neglecting the interpretation of their results. This leads to a considerable lack of transparency, posing a significant risk in the actual diagnostic process. To solve this problem, this paper introduces MambaCapsule, a deep neural networks for ECG arrhythmias classification, which increases the explainability of the model while enhancing the accuracy.Our model utilizes Mamba for feature extraction and Capsule networks for prediction, providing not only a confidence score but also signal features. Akin to the processing mechanism of human brain, the model learns signal features and their relationship between them by reconstructing ECG signals in the predicted selection. The model evaluation was conducted on MIT-BIH and PTB dataset, following the AAMI standard. MambaCapsule has achieved a total accuracy of 99.54% and 99.59% on the test sets respectively. These results demonstrate the promising performance of under the standard test protocol.
TWIN-GPT: Digital Twins for Clinical Trials via Large Language Model
Apr 01, 2024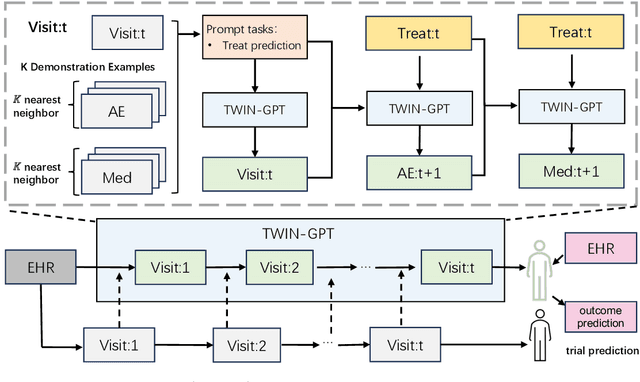


Abstract:Recently, there has been a burgeoning interest in virtual clinical trials, which simulate real-world scenarios and hold the potential to significantly enhance patient safety, expedite development, reduce costs, and contribute to the broader scientific knowledge in healthcare. Existing research often focuses on leveraging electronic health records (EHRs) to support clinical trial outcome prediction. Yet, trained with limited clinical trial outcome data, existing approaches frequently struggle to perform accurate predictions. Some research has attempted to generate EHRs to augment model development but has fallen short in personalizing the generation for individual patient profiles. Recently, the emergence of large language models has illuminated new possibilities, as their embedded comprehensive clinical knowledge has proven beneficial in addressing medical issues. In this paper, we propose a large language model-based digital twin creation approach, called TWIN-GPT. TWIN-GPT can establish cross-dataset associations of medical information given limited data, generating unique personalized digital twins for different patients, thereby preserving individual patient characteristics. Comprehensive experiments show that using digital twins created by TWIN-GPT can boost clinical trial outcome prediction, exceeding various previous prediction approaches. Besides, we also demonstrate that TWIN-GPT can generate high-fidelity trial data that closely approximate specific patients, aiding in more accurate result predictions in data-scarce situations. Moreover, our study provides practical evidence for the application of digital twins in healthcare, highlighting its potential significance.
D-Former: A U-shaped Dilated Transformer for 3D Medical Image Segmentation
Jan 10, 2022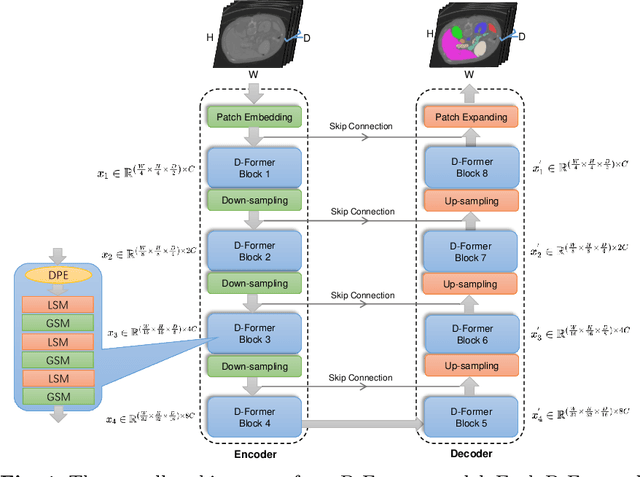
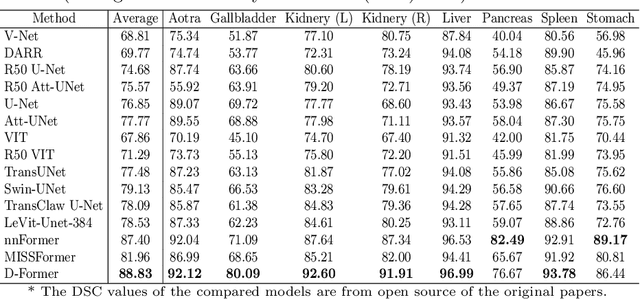
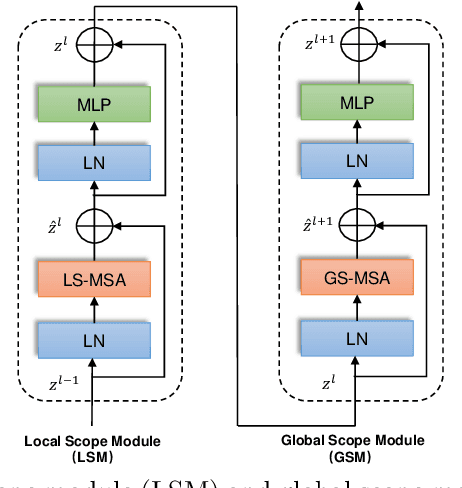
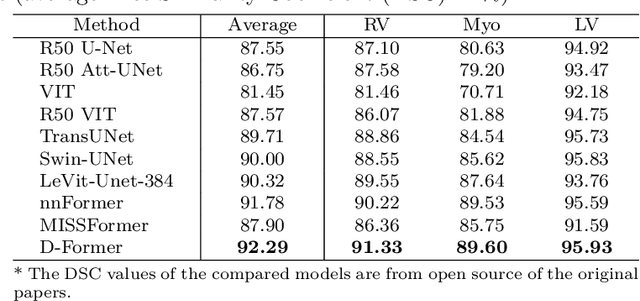
Abstract:Computer-aided medical image segmentation has been applied widely in diagnosis and treatment to obtain clinically useful information of shapes and volumes of target organs and tissues. In the past several years, convolutional neural network (CNN) based methods (e.g., U-Net) have dominated this area, but still suffered from inadequate long-range information capturing. Hence, recent work presented computer vision Transformer variants for medical image segmentation tasks and obtained promising performances. Such Transformers model long-range dependency by computing pair-wise patch relations. However, they incur prohibitive computational costs, especially on 3D medical images (e.g., CT and MRI). In this paper, we propose a new method called Dilated Transformer, which conducts self-attention for pair-wise patch relations captured alternately in local and global scopes. Inspired by dilated convolution kernels, we conduct the global self-attention in a dilated manner, enlarging receptive fields without increasing the patches involved and thus reducing computational costs. Based on this design of Dilated Transformer, we construct a U-shaped encoder-decoder hierarchical architecture called D-Former for 3D medical image segmentation. Experiments on the Synapse and ACDC datasets show that our D-Former model, trained from scratch, outperforms various competitive CNN-based or Transformer-based segmentation models at a low computational cost without time-consuming per-training process.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge