Kuanlun Liao
ME-GAN: Learning Panoptic Electrocardio Representations for Multi-view ECG Synthesis Conditioned on Heart Diseases
Jul 21, 2022



Abstract:Electrocardiogram (ECG) is a widely used non-invasive diagnostic tool for heart diseases. Many studies have devised ECG analysis models (e.g., classifiers) to assist diagnosis. As an upstream task, researches have built generative models to synthesize ECG data, which are beneficial to providing training samples, privacy protection, and annotation reduction. However, previous generative methods for ECG often neither synthesized multi-view data, nor dealt with heart disease conditions. In this paper, we propose a novel disease-aware generative adversarial network for multi-view ECG synthesis called ME-GAN, which attains panoptic electrocardio representations conditioned on heart diseases and projects the representations onto multiple standard views to yield ECG signals. Since ECG manifestations of heart diseases are often localized in specific waveforms, we propose a new "mixup normalization" to inject disease information precisely into suitable locations. In addition, we propose a view discriminator to revert disordered ECG views into a pre-determined order, supervising the generator to obtain ECG representing correct view characteristics. Besides, a new metric, rFID, is presented to assess the quality of the synthesized ECG signals. Comprehensive experiments verify that our ME-GAN performs well on multi-view ECG signal synthesis with trusty morbid manifestations.
D-Former: A U-shaped Dilated Transformer for 3D Medical Image Segmentation
Jan 10, 2022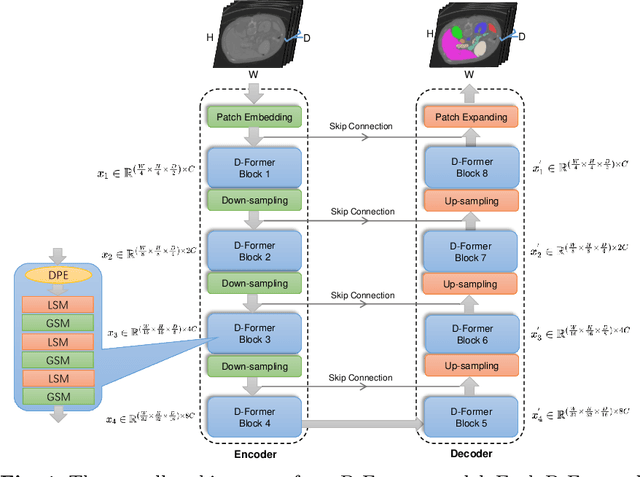
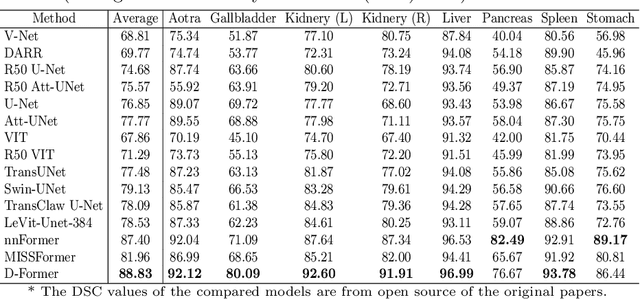
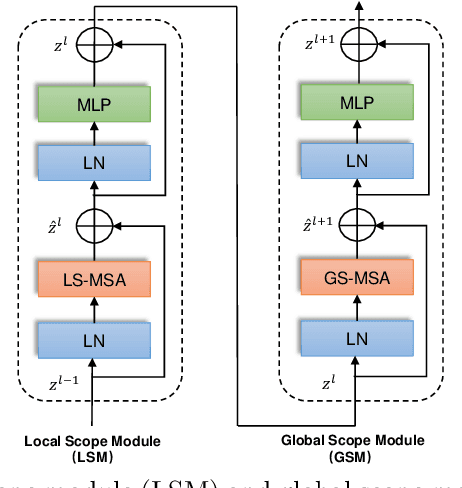
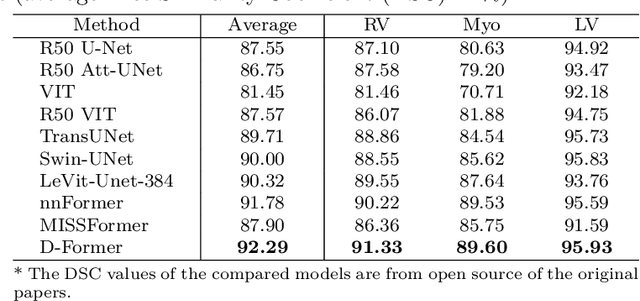
Abstract:Computer-aided medical image segmentation has been applied widely in diagnosis and treatment to obtain clinically useful information of shapes and volumes of target organs and tissues. In the past several years, convolutional neural network (CNN) based methods (e.g., U-Net) have dominated this area, but still suffered from inadequate long-range information capturing. Hence, recent work presented computer vision Transformer variants for medical image segmentation tasks and obtained promising performances. Such Transformers model long-range dependency by computing pair-wise patch relations. However, they incur prohibitive computational costs, especially on 3D medical images (e.g., CT and MRI). In this paper, we propose a new method called Dilated Transformer, which conducts self-attention for pair-wise patch relations captured alternately in local and global scopes. Inspired by dilated convolution kernels, we conduct the global self-attention in a dilated manner, enlarging receptive fields without increasing the patches involved and thus reducing computational costs. Based on this design of Dilated Transformer, we construct a U-shaped encoder-decoder hierarchical architecture called D-Former for 3D medical image segmentation. Experiments on the Synapse and ACDC datasets show that our D-Former model, trained from scratch, outperforms various competitive CNN-based or Transformer-based segmentation models at a low computational cost without time-consuming per-training process.
DANets: Deep Abstract Networks for Tabular Data Classification and Regression
Dec 24, 2021



Abstract:Tabular data are ubiquitous in real world applications. Although many commonly-used neural components (e.g., convolution) and extensible neural networks (e.g., ResNet) have been developed by the machine learning community, few of them were effective for tabular data and few designs were adequately tailored for tabular data structures. In this paper, we propose a novel and flexible neural component for tabular data, called Abstract Layer (AbstLay), which learns to explicitly group correlative input features and generate higher-level features for semantics abstraction. Also, we design a structure re-parameterization method to compress AbstLay, thus reducing the computational complexity by a clear margin in the reference phase. A special basic block is built using AbstLays, and we construct a family of Deep Abstract Networks (DANets) for tabular data classification and regression by stacking such blocks. In DANets, a special shortcut path is introduced to fetch information from raw tabular features, assisting feature interactions across different levels. Comprehensive experiments on seven real-world tabular datasets show that our AbstLay and DANets are effective for tabular data classification and regression, and the computational complexity is superior to competitive methods. Besides, we evaluate the performance gains of DANet as it goes deep, verifying the extendibility of our method. Our code is available at https://github.com/WhatAShot/DANet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge