Danny Z. Chen
H-CNN-ViT: A Hierarchical Gated Attention Multi-Branch Model for Bladder Cancer Recurrence Prediction
Nov 19, 2025Abstract:Bladder cancer is one of the most prevalent malignancies worldwide, with a recurrence rate of up to 78%, necessitating accurate post-operative monitoring for effective patient management. Multi-sequence contrast-enhanced MRI is commonly used for recurrence detection; however, interpreting these scans remains challenging, even for experienced radiologists, due to post-surgical alterations such as scarring, swelling, and tissue remodeling. AI-assisted diagnostic tools have shown promise in improving bladder cancer recurrence prediction, yet progress in this field is hindered by the lack of dedicated multi-sequence MRI datasets for recurrence assessment study. In this work, we first introduce a curated multi-sequence, multi-modal MRI dataset specifically designed for bladder cancer recurrence prediction, establishing a valuable benchmark for future research. We then propose H-CNN-ViT, a new Hierarchical Gated Attention Multi-Branch model that enables selective weighting of features from the global (ViT) and local (CNN) paths based on contextual demands, achieving a balanced and targeted feature fusion. Our multi-branch architecture processes each modality independently, ensuring that the unique properties of each imaging channel are optimally captured and integrated. Evaluated on our dataset, H-CNN-ViT achieves an AUC of 78.6%, surpassing state-of-the-art models. Our model is publicly available at https://github.com/XLIAaron/H-CNN-ViT.
Versatile and Risk-Sensitive Cardiac Diagnosis via Graph-Based ECG Signal Representation
Nov 11, 2025Abstract:Despite the rapid advancements of electrocardiogram (ECG) signal diagnosis and analysis methods through deep learning, two major hurdles still limit their clinical adoption: the lack of versatility in processing ECG signals with diverse configurations, and the inadequate detection of risk signals due to sample imbalances. Addressing these challenges, we introduce VersAtile and Risk-Sensitive cardiac diagnosis (VARS), an innovative approach that employs a graph-based representation to uniformly model heterogeneous ECG signals. VARS stands out by transforming ECG signals into versatile graph structures that capture critical diagnostic features, irrespective of signal diversity in the lead count, sampling frequency, and duration. This graph-centric formulation also enhances diagnostic sensitivity, enabling precise localization and identification of abnormal ECG patterns that often elude standard analysis methods. To facilitate representation transformation, our approach integrates denoising reconstruction with contrastive learning to preserve raw ECG information while highlighting pathognomonic patterns. We rigorously evaluate the efficacy of VARS on three distinct ECG datasets, encompassing a range of structural variations. The results demonstrate that VARS not only consistently surpasses existing state-of-the-art models across all these datasets but also exhibits substantial improvement in identifying risk signals. Additionally, VARS offers interpretability by pinpointing the exact waveforms that lead to specific model outputs, thereby assisting clinicians in making informed decisions. These findings suggest that our VARS will likely emerge as an invaluable tool for comprehensive cardiac health assessment.
When Swin Transformer Meets KANs: An Improved Transformer Architecture for Medical Image Segmentation
Nov 06, 2025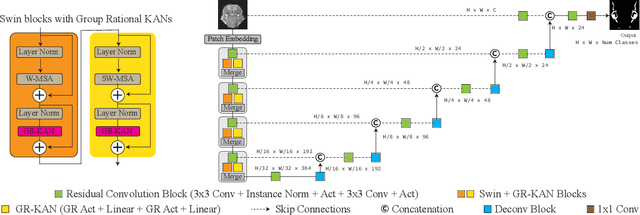
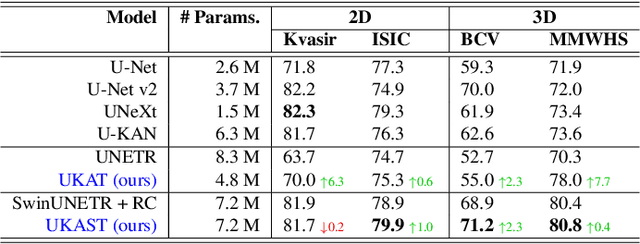
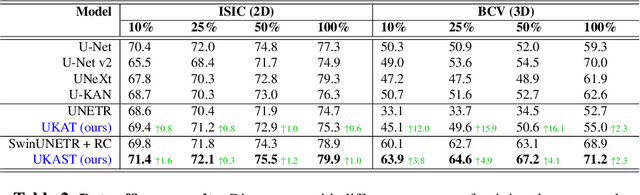
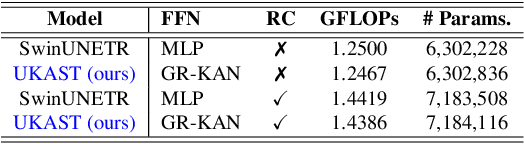
Abstract:Medical image segmentation is critical for accurate diagnostics and treatment planning, but remains challenging due to complex anatomical structures and limited annotated training data. CNN-based segmentation methods excel at local feature extraction, but struggle with modeling long-range dependencies. Transformers, on the other hand, capture global context more effectively, but are inherently data-hungry and computationally expensive. In this work, we introduce UKAST, a U-Net like architecture that integrates rational-function based Kolmogorov-Arnold Networks (KANs) into Swin Transformer encoders. By leveraging rational base functions and Group Rational KANs (GR-KANs) from the Kolmogorov-Arnold Transformer (KAT), our architecture addresses the inefficiencies of vanilla spline-based KANs, yielding a more expressive and data-efficient framework with reduced FLOPs and only a very small increase in parameter count compared to SwinUNETR. UKAST achieves state-of-the-art performance on four diverse 2D and 3D medical image segmentation benchmarks, consistently surpassing both CNN- and Transformer-based baselines. Notably, it attains superior accuracy in data-scarce settings, alleviating the data-hungry limitations of standard Vision Transformers. These results show the potential of KAN-enhanced Transformers to advance data-efficient medical image segmentation. Code is available at: https://github.com/nsapkota417/UKAST
Adapting a Segmentation Foundation Model for Medical Image Classification
May 09, 2025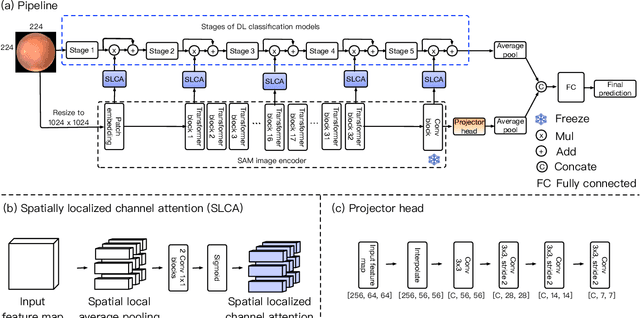
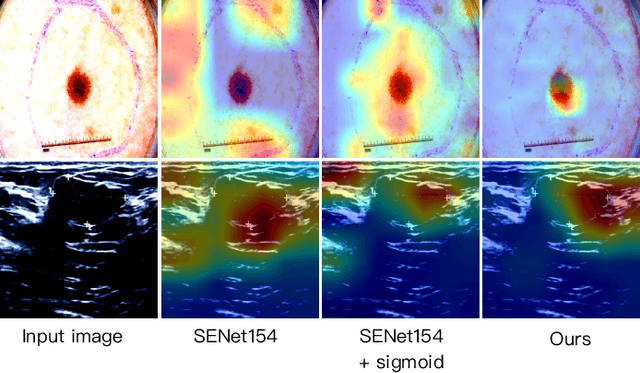
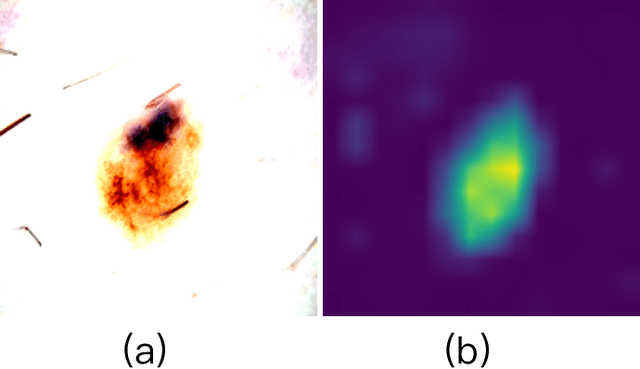
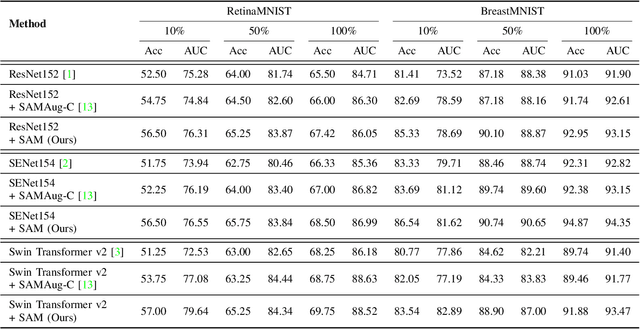
Abstract:Recent advancements in foundation models, such as the Segment Anything Model (SAM), have shown strong performance in various vision tasks, particularly image segmentation, due to their impressive zero-shot segmentation capabilities. However, effectively adapting such models for medical image classification is still a less explored topic. In this paper, we introduce a new framework to adapt SAM for medical image classification. First, we utilize the SAM image encoder as a feature extractor to capture segmentation-based features that convey important spatial and contextual details of the image, while freezing its weights to avoid unnecessary overhead during training. Next, we propose a novel Spatially Localized Channel Attention (SLCA) mechanism to compute spatially localized attention weights for the feature maps. The features extracted from SAM's image encoder are processed through SLCA to compute attention weights, which are then integrated into deep learning classification models to enhance their focus on spatially relevant or meaningful regions of the image, thus improving classification performance. Experimental results on three public medical image classification datasets demonstrate the effectiveness and data-efficiency of our approach.
A Survey on Ordinal Regression: Applications, Advances and Prospects
Mar 02, 2025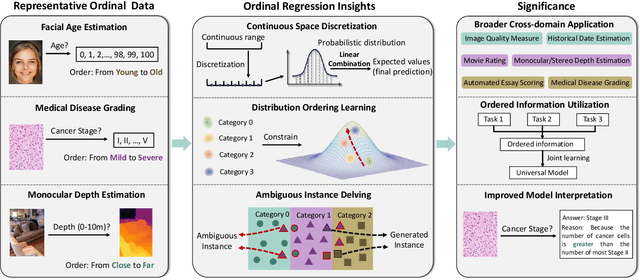
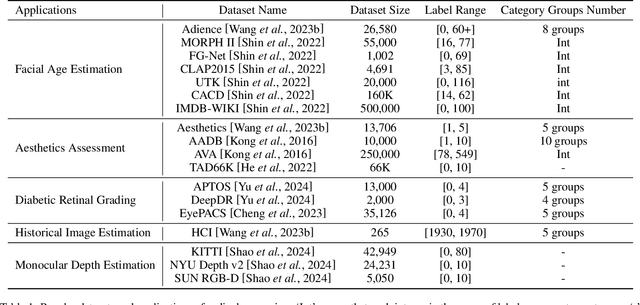
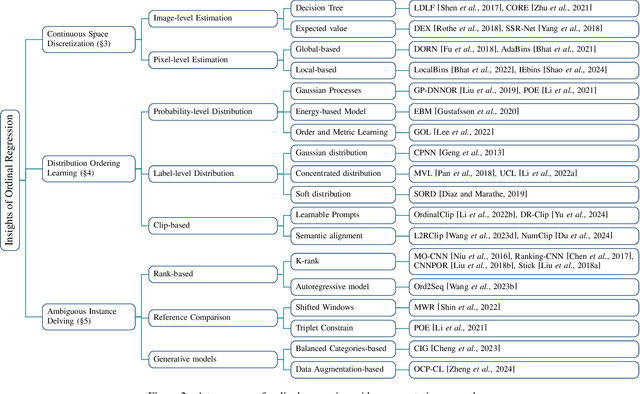
Abstract:Ordinal regression refers to classifying object instances into ordinal categories. Ordinal regression is crucial for applications in various areas like facial age estimation, image aesthetics assessment, and even cancer staging, due to its capability to utilize ordered information effectively. More importantly, it also enhances model interpretation by considering category order, aiding the understanding of data trends and causal relationships. Despite significant recent progress, challenges remain, and further investigation of ordinal regression techniques and applications is essential to guide future research. In this survey, we present a comprehensive examination of advances and applications of ordinal regression. By introducing a systematic taxonomy, we meticulously classify the pertinent techniques and applications into three well-defined categories based on different strategies and objectives: Continuous Space Discretization, Distribution Ordering Learning, and Ambiguous Instance Delving. This categorization enables a structured exploration of diverse insights in ordinal regression problems, providing a framework for a more comprehensive understanding and evaluation of this field and its related applications. To our best knowledge, this is the first systematic survey of ordinal regression, which lays a foundation for future research in this fundamental and generic domain.
Sli2Vol+: Segmenting 3D Medical Images Based on an Object Estimation Guided Correspondence Flow Network
Nov 21, 2024



Abstract:Deep learning (DL) methods have shown remarkable successes in medical image segmentation, often using large amounts of annotated data for model training. However, acquiring a large number of diverse labeled 3D medical image datasets is highly difficult and expensive. Recently, mask propagation DL methods were developed to reduce the annotation burden on 3D medical images. For example, Sli2Vol~\cite{yeung2021sli2vol} proposed a self-supervised framework (SSF) to learn correspondences by matching neighboring slices via slice reconstruction in the training stage; the learned correspondences were then used to propagate a labeled slice to other slices in the test stage. But, these methods are still prone to error accumulation due to the inter-slice propagation of reconstruction errors. Also, they do not handle discontinuities well, which can occur between consecutive slices in 3D images, as they emphasize exploiting object continuity. To address these challenges, in this work, we propose a new SSF, called \proposed, {for segmenting any anatomical structures in 3D medical images using only a single annotated slice per training and testing volume.} Specifically, in the training stage, we first propagate an annotated 2D slice of a training volume to the other slices, generating pseudo-labels (PLs). Then, we develop a novel Object Estimation Guided Correspondence Flow Network to learn reliable correspondences between consecutive slices and corresponding PLs in a self-supervised manner. In the test stage, such correspondences are utilized to propagate a single annotated slice to the other slices of a test volume. We demonstrate the effectiveness of our method on various medical image segmentation tasks with different datasets, showing better generalizability across different organs, modalities, and modals. Code is available at \url{https://github.com/adlsn/Sli2Volplus}
UniCoN: Universal Conditional Networks for Multi-Age Embryonic Cartilage Segmentation with Sparsely Annotated Data
Oct 16, 2024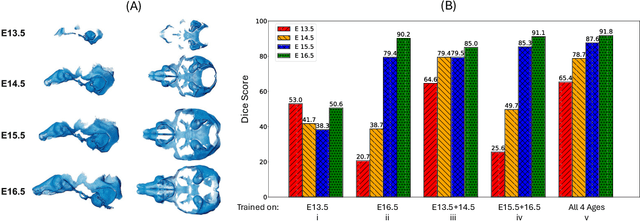
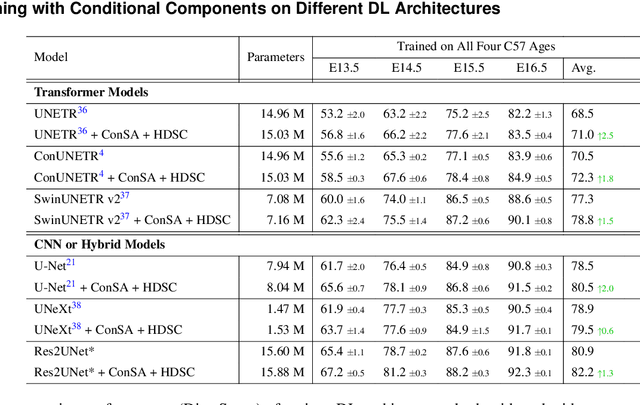
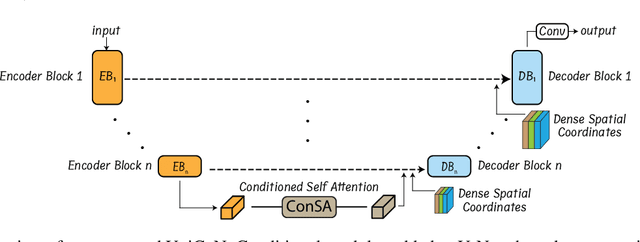
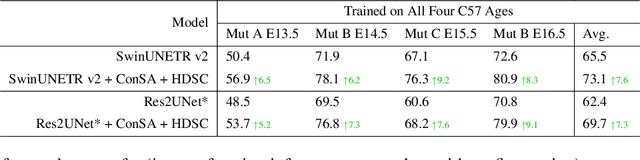
Abstract:Osteochondrodysplasia, affecting 2-3% of newborns globally, is a group of bone and cartilage disorders that often result in head malformations, contributing to childhood morbidity and reduced quality of life. Current research on this disease using mouse models faces challenges since it involves accurately segmenting the developing cartilage in 3D micro-CT images of embryonic mice. Tackling this segmentation task with deep learning (DL) methods is laborious due to the big burden of manual image annotation, expensive due to the high acquisition costs of 3D micro-CT images, and difficult due to embryonic cartilage's complex and rapidly changing shapes. While DL approaches have been proposed to automate cartilage segmentation, most such models have limited accuracy and generalizability, especially across data from different embryonic age groups. To address these limitations, we propose novel DL methods that can be adopted by any DL architectures -- including CNNs, Transformers, or hybrid models -- which effectively leverage age and spatial information to enhance model performance. Specifically, we propose two new mechanisms, one conditioned on discrete age categories and the other on continuous image crop locations, to enable an accurate representation of cartilage shape changes across ages and local shape details throughout the cranial region. Extensive experiments on multi-age cartilage segmentation datasets show significant and consistent performance improvements when integrating our conditional modules into popular DL segmentation architectures. On average, we achieve a 1.7% Dice score increase with minimal computational overhead and a 7.5% improvement on unseen data. These results highlight the potential of our approach for developing robust, universal models capable of handling diverse datasets with limited annotated data, a key challenge in DL-based medical image analysis.
FiAt-Net: Detecting Fibroatheroma Plaque Cap in 3D Intravascular OCT Images
Sep 13, 2024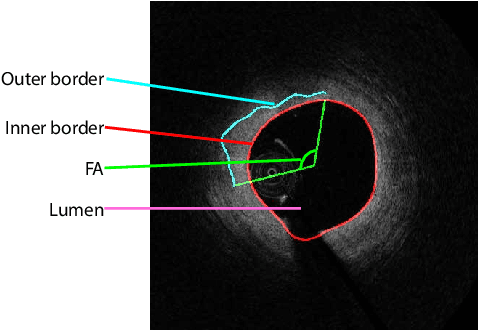
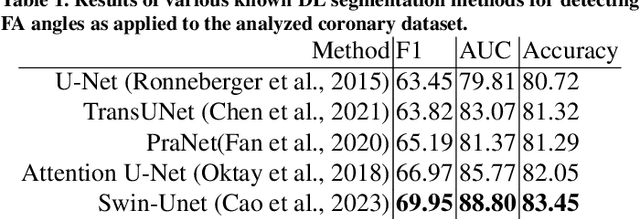
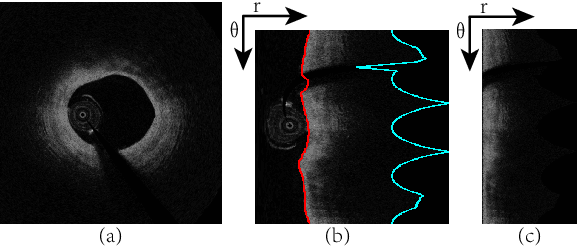
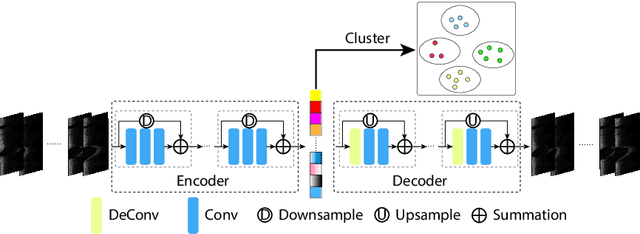
Abstract:The key manifestation of coronary artery disease (CAD) is development of fibroatheromatous plaque, the cap of which may rupture and subsequently lead to coronary artery blocking and heart attack. As such, quantitative analysis of coronary plaque, its plaque cap, and consequently the cap's likelihood to rupture are of critical importance when assessing a risk of cardiovascular events. This paper reports a new deep learning based approach, called FiAt-Net, for detecting angular extent of fibroatheroma (FA) and segmenting its cap in 3D intravascular optical coherence tomography (IVOCT) images. IVOCT 2D image frames are first associated with distinct clusters and data from each cluster are used for model training. As plaque is typically focal and thus unevenly distributed, a binary partitioning method is employed to identify FA plaque areas to focus on to mitigate the data imbalance issue. Additional image representations (called auxiliary images) are generated to capture IVOCT intensity changes to help distinguish FA and non-FA areas on the coronary wall. Information in varying scales is derived from the original IVOCT and auxiliary images, and a multi-head self-attention mechanism is employed to fuse such information. Our FiAt-Net achieved high performance on a 3D IVOCT coronary image dataset, demonstrating its effectiveness in accurately detecting FA cap in IVOCT images.
Spectral U-Net: Enhancing Medical Image Segmentation via Spectral Decomposition
Sep 13, 2024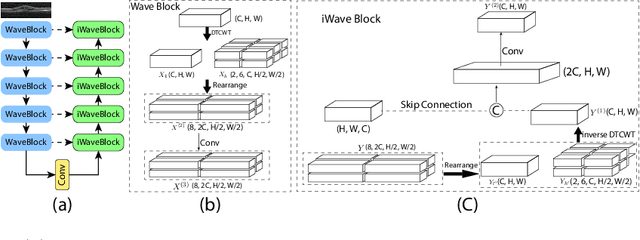

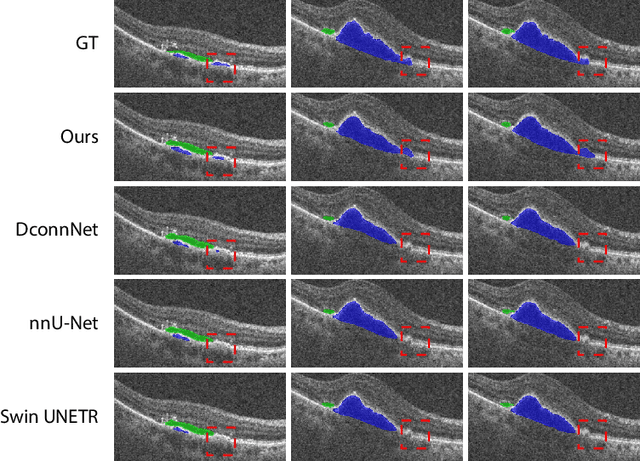

Abstract:This paper introduces Spectral U-Net, a novel deep learning network based on spectral decomposition, by exploiting Dual Tree Complex Wavelet Transform (DTCWT) for down-sampling and inverse Dual Tree Complex Wavelet Transform (iDTCWT) for up-sampling. We devise the corresponding Wave-Block and iWave-Block, integrated into the U-Net architecture, aiming at mitigating information loss during down-sampling and enhancing detail reconstruction during up-sampling. In the encoder, we first decompose the feature map into high and low-frequency components using DTCWT, enabling down-sampling while mitigating information loss. In the decoder, we utilize iDTCWT to reconstruct higher-resolution feature maps from down-sampled features. Evaluations on the Retina Fluid, Brain Tumor, and Liver Tumor segmentation datasets with the nnU-Net framework demonstrate the superiority of the proposed Spectral U-Net.
TeleOR: Real-time Telemedicine System for Full-Scene Operating Room
Jul 29, 2024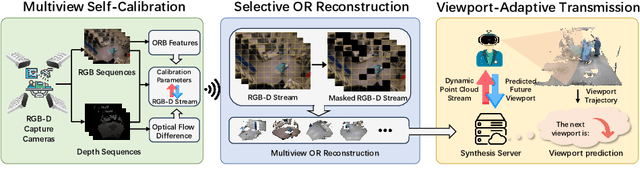

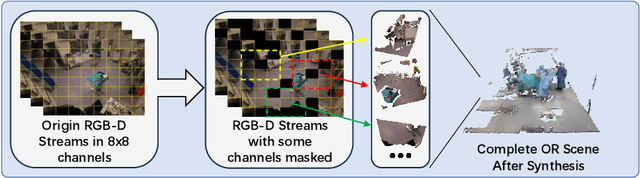
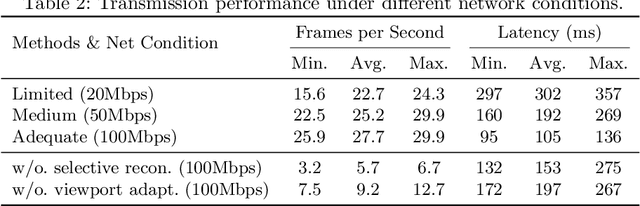
Abstract:The advent of telemedicine represents a transformative development in leveraging technology to extend the reach of specialized medical expertise to remote surgeries, a field where the immediacy of expert guidance is paramount. However, the intricate dynamics of Operating Room (OR) scene pose unique challenges for telemedicine, particularly in achieving high-fidelity, real-time scene reconstruction and transmission amidst obstructions and bandwidth limitations. This paper introduces TeleOR, a pioneering system designed to address these challenges through real-time OR scene reconstruction for Tele-intervention. TeleOR distinguishes itself with three innovative approaches: dynamic self-calibration, which leverages inherent scene features for calibration without the need for preset markers, allowing for obstacle avoidance and real-time camera adjustment; selective OR reconstruction, focusing on dynamically changing scene segments to reduce reconstruction complexity; and viewport-adaptive transmission, optimizing data transmission based on real-time client feedback to efficiently deliver high-quality 3D reconstructions within bandwidth constraints. Comprehensive experiments on the 4D-OR surgical scene dataset demostrate the superiority and applicability of TeleOR, illuminating the potential to revolutionize tele-interventions by overcoming the spatial and technical barriers inherent in remote surgical guidance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge