Hongxiao Wang
Toward Auditable Neuro-Symbolic Reasoning in Pathology: SQL as an Explicit Trace of Evidence
Jan 05, 2026Abstract:Automated pathology image analysis is central to clinical diagnosis, but clinicians still ask which slide features drive a model's decision and why. Vision-language models can produce natural language explanations, but these are often correlational and lack verifiable evidence. In this paper, we introduce an SQL-centered agentic framework that enables both feature measurement and reasoning to be auditable. Specifically, after extracting human-interpretable cellular features, Feature Reasoning Agents compose and execute SQL queries over feature tables to aggregate visual evidence into quantitative findings. A Knowledge Comparison Agent then evaluates these findings against established pathological knowledge, mirroring how pathologists justify diagnoses from measurable observations. Extensive experiments evaluated on two pathology visual question answering datasets demonstrate our method improves interpretability and decision traceability while producing executable SQL traces that link cellular measurements to diagnostic conclusions.
FlowRoI A Fast Optical Flow Driven Region of Interest Extraction Framework for High-Throughput Image Compression in Immune Cell Migration Analysis
Nov 18, 2025Abstract:Autonomous migration is essential for the function of immune cells such as neutrophils and plays a pivotal role in diverse diseases. Recently, we introduced ComplexEye, a multi-lens array microscope comprising 16 independent aberration-corrected glass lenses arranged at the pitch of a 96-well plate, capable of capturing high-resolution movies of migrating cells. This architecture enables high-throughput live-cell video microscopy for migration analysis, supporting routine quantification of autonomous motility with strong potential for clinical translation. However, ComplexEye and similar high-throughput imaging platforms generate data at an exponential rate, imposing substantial burdens on storage and transmission. To address this challenge, we present FlowRoI, a fast optical-flow-based region of interest (RoI) extraction framework designed for high-throughput image compression in immune cell migration studies. FlowRoI estimates optical flow between consecutive frames and derives RoI masks that reliably cover nearly all migrating cells. The raw image and its corresponding RoI mask are then jointly encoded using JPEG2000 to enable RoI-aware compression. FlowRoI operates with high computational efficiency, achieving runtimes comparable to standard JPEG2000 and reaching an average throughput of about 30 frames per second on a modern laptop equipped with an Intel i7-1255U CPU. In terms of image quality, FlowRoI yields higher peak signal-to-noise ratio (PSNR) in cellular regions and achieves 2.0-2.2x higher compression rates at matched PSNR compared to standard JPEG2000.
PathMR: Multimodal Visual Reasoning for Interpretable Pathology Diagnosis
Aug 28, 2025Abstract:Deep learning based automated pathological diagnosis has markedly improved diagnostic efficiency and reduced variability between observers, yet its clinical adoption remains limited by opaque model decisions and a lack of traceable rationale. To address this, recent multimodal visual reasoning architectures provide a unified framework that generates segmentation masks at the pixel level alongside semantically aligned textual explanations. By localizing lesion regions and producing expert style diagnostic narratives, these models deliver the transparent and interpretable insights necessary for dependable AI assisted pathology. Building on these advancements, we propose PathMR, a cell-level Multimodal visual Reasoning framework for Pathological image analysis. Given a pathological image and a textual query, PathMR generates expert-level diagnostic explanations while simultaneously predicting cell distribution patterns. To benchmark its performance, we evaluated our approach on the publicly available PathGen dataset as well as on our newly developed GADVR dataset. Extensive experiments on these two datasets demonstrate that PathMR consistently outperforms state-of-the-art visual reasoning methods in text generation quality, segmentation accuracy, and cross-modal alignment. These results highlight the potential of PathMR for improving interpretability in AI-driven pathological diagnosis. The code will be publicly available in https://github.com/zhangye-zoe/PathMR.
Diff-CL: A Novel Cross Pseudo-Supervision Method for Semi-supervised Medical Image Segmentation
Mar 12, 2025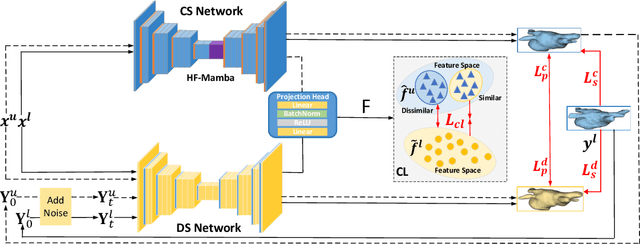
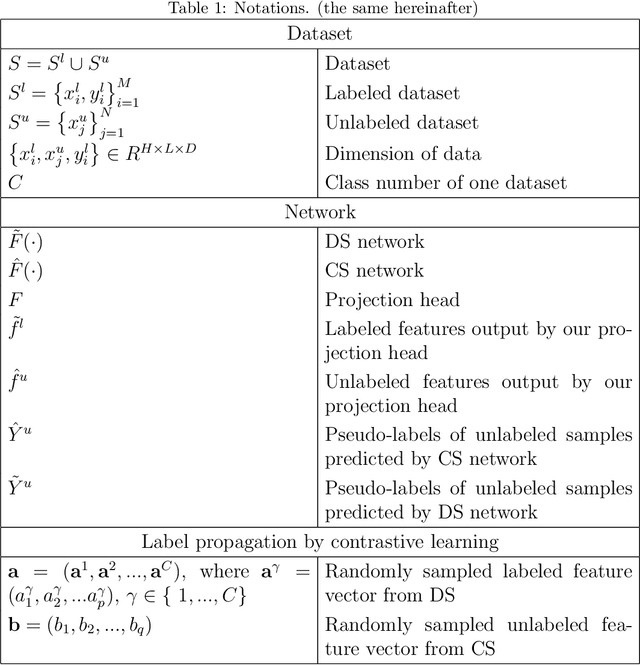
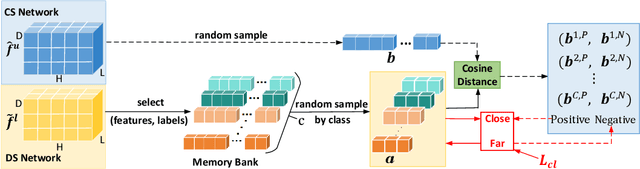
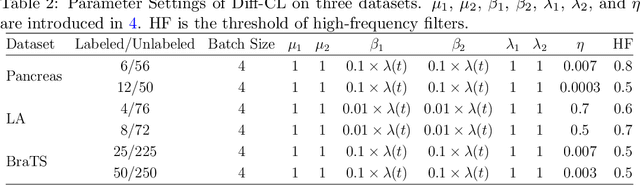
Abstract:Semi-supervised learning utilizes insights from unlabeled data to improve model generalization, thereby reducing reliance on large labeled datasets. Most existing studies focus on limited samples and fail to capture the overall data distribution. We contend that combining distributional information with detailed information is crucial for achieving more robust and accurate segmentation results. On the one hand, with its robust generative capabilities, diffusion models (DM) learn data distribution effectively. However, it struggles with fine detail capture, leading to generated images with misleading details. Combining DM with convolutional neural networks (CNNs) enables the former to learn data distribution while the latter corrects fine details. While capturing complete high-frequency details by CNNs requires substantial computational resources and is susceptible to local noise. On the other hand, given that both labeled and unlabeled data come from the same distribution, we believe that regions in unlabeled data similar to overall class semantics to labeled data are likely to belong to the same class, while regions with minimal similarity are less likely to. This work introduces a semi-supervised medical image segmentation framework from the distribution perspective (Diff-CL). Firstly, we propose a cross-pseudo-supervision learning mechanism between diffusion and convolution segmentation networks. Secondly, we design a high-frequency mamba module to capture boundary and detail information globally. Finally, we apply contrastive learning for label propagation from labeled to unlabeled data. Our method achieves state-of-the-art (SOTA) performance across three datasets, including left atrium, brain tumor, and NIH pancreas datasets.
GIGP: A Global Information Interacting and Geometric Priors Focusing Framework for Semi-supervised Medical Image Segmentation
Mar 12, 2025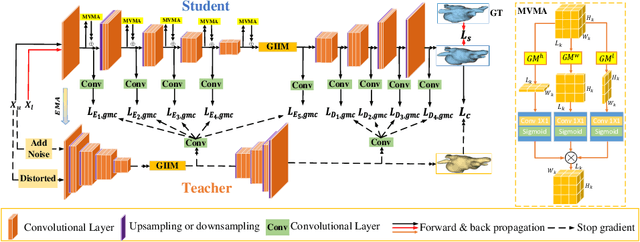
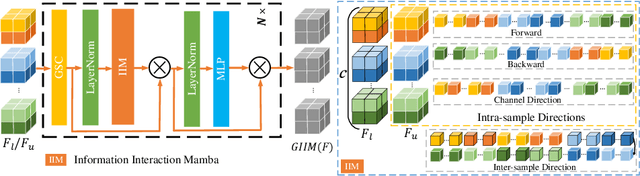
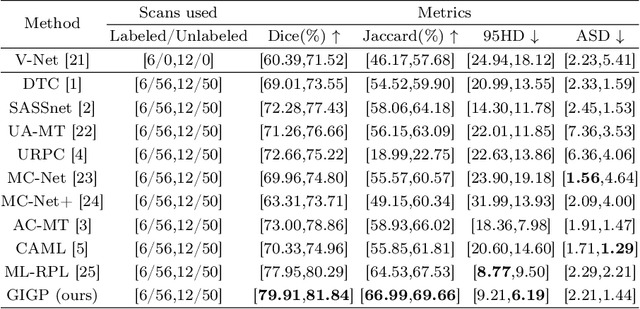
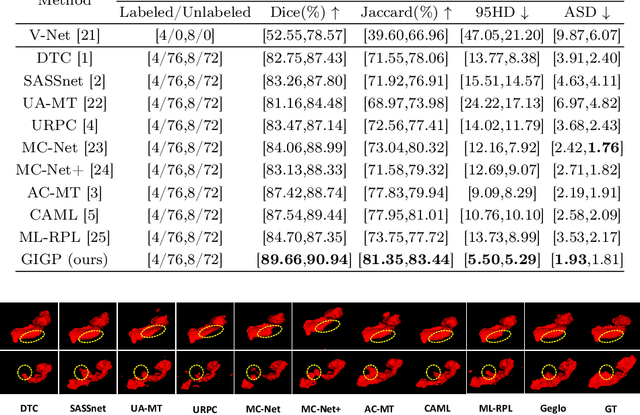
Abstract:Semi-supervised learning enhances medical image segmentation by leveraging unlabeled data, reducing reliance on extensive labeled datasets. On the one hand, the distribution discrepancy between limited labeled data and abundant unlabeled data can hinder model generalization. Most existing methods rely on local similarity matching, which may introduce bias. In contrast, Mamba effectively models global context with linear complexity, learning more comprehensive data representations. On the other hand, medical images usually exhibit consistent anatomical structures defined by geometric features. Most existing methods fail to fully utilize global geometric priors, such as volumes, moments etc. In this work, we introduce a global information interaction and geometric priors focus framework (GIGP). Firstly, we present a Global Information Interaction Mamba module to reduce distribution discrepancy between labeled and unlabeled data. Secondly, we propose a Geometric Moment Attention Mechanism to extract richer global geometric features. Finally, we propose Global Geometric Perturbation Consistency to simulate organ dynamics and geometric variations, enhancing the ability of the model to learn generalized features. The superior performance on the NIH Pancreas and Left Atrium datasets demonstrates the effectiveness of our approach.
Boosting Medical Image Classification with Segmentation Foundation Model
Jun 16, 2024
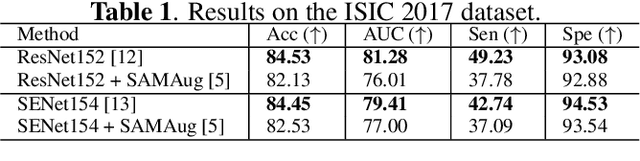
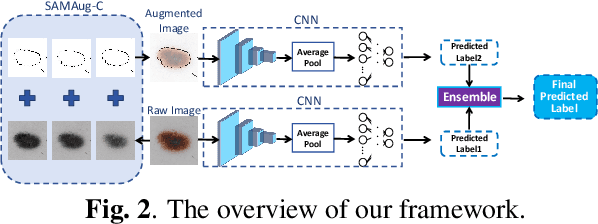
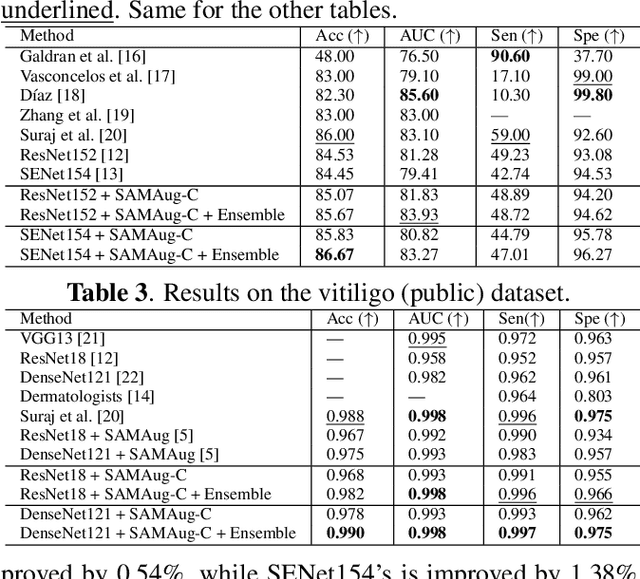
Abstract:The Segment Anything Model (SAM) exhibits impressive capabilities in zero-shot segmentation for natural images. Recently, SAM has gained a great deal of attention for its applications in medical image segmentation. However, to our best knowledge, no studies have shown how to harness the power of SAM for medical image classification. To fill this gap and make SAM a true ``foundation model'' for medical image analysis, it is highly desirable to customize SAM specifically for medical image classification. In this paper, we introduce SAMAug-C, an innovative augmentation method based on SAM for augmenting classification datasets by generating variants of the original images. The augmented datasets can be used to train a deep learning classification model, thereby boosting the classification performance. Furthermore, we propose a novel framework that simultaneously processes raw and SAMAug-C augmented image input, capitalizing on the complementary information that is offered by both. Experiments on three public datasets validate the effectiveness of our new approach.
Self Pre-training with Topology- and Spatiality-aware Masked Autoencoders for 3D Medical Image Segmentation
Jun 15, 2024
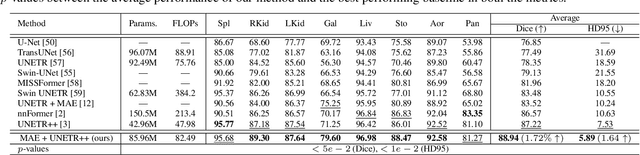
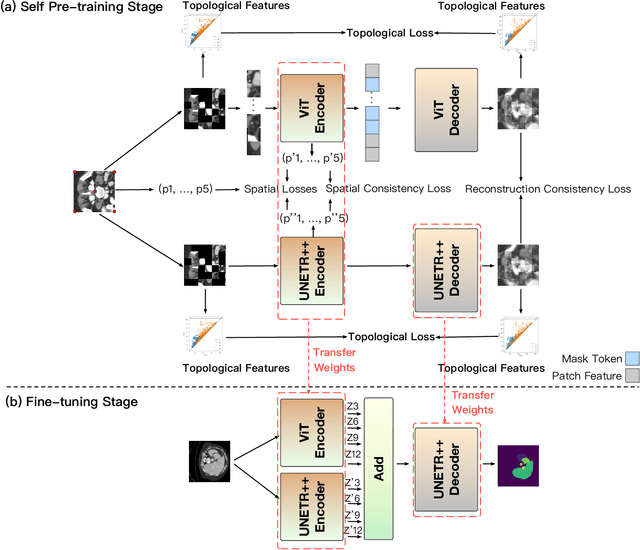
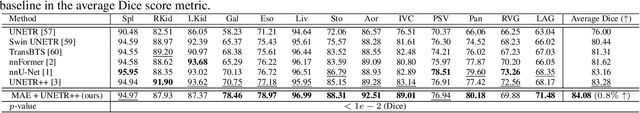
Abstract:Masked Autoencoders (MAEs) have been shown to be effective in pre-training Vision Transformers (ViTs) for natural and medical image analysis problems. By reconstructing missing pixel/voxel information in visible patches, a ViT encoder can aggregate contextual information for downstream tasks. But, existing MAE pre-training methods, which were specifically developed with the ViT architecture, lack the ability to capture geometric shape and spatial information, which is critical for medical image segmentation tasks. In this paper, we propose a novel extension of known MAEs for self pre-training (i.e., models pre-trained on the same target dataset) for 3D medical image segmentation. (1) We propose a new topological loss to preserve geometric shape information by computing topological signatures of both the input and reconstructed volumes, learning geometric shape information. (2) We introduce a pre-text task that predicts the positions of the centers and eight corners of 3D crops, enabling the MAE to aggregate spatial information. (3) We extend the MAE pre-training strategy to a hybrid state-of-the-art (SOTA) medical image segmentation architecture and co-pretrain it alongside the ViT. (4) We develop a fine-tuned model for downstream segmentation tasks by complementing the pre-trained ViT encoder with our pre-trained SOTA model. Extensive experiments on five public 3D segmentation datasets show the effectiveness of our new approach.
Path-GPTOmic: A Balanced Multi-modal Learning Framework for Survival Outcome Prediction
Mar 18, 2024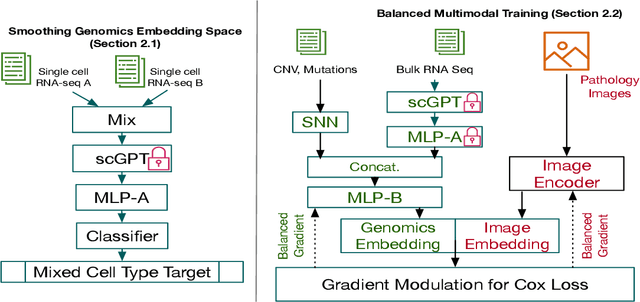
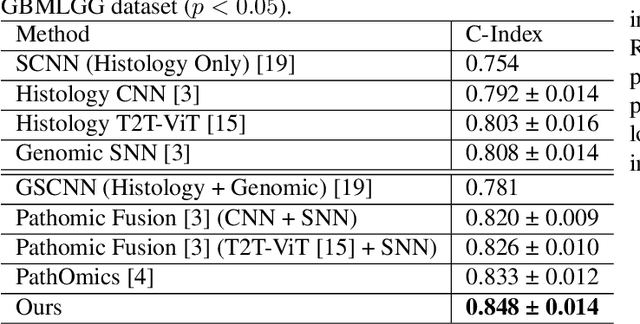

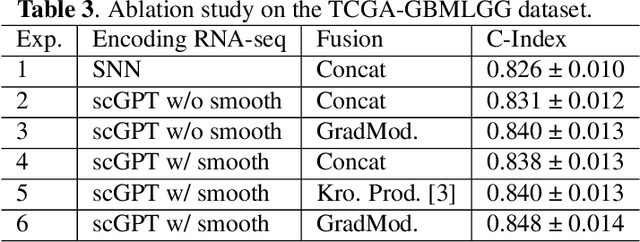
Abstract:For predicting cancer survival outcomes, standard approaches in clinical research are often based on two main modalities: pathology images for observing cell morphology features, and genomic (e.g., bulk RNA-seq) for quantifying gene expressions. However, existing pathology-genomic multi-modal algorithms face significant challenges: (1) Valuable biological insights regarding genes and gene-gene interactions are frequently overlooked; (2) one modality often dominates the optimization process, causing inadequate training for the other modality. In this paper, we introduce a new multi-modal ``Path-GPTOmic" framework for cancer survival outcome prediction. First, to extract valuable biological insights, we regulate the embedding space of a foundation model, scGPT, initially trained on single-cell RNA-seq data, making it adaptable for bulk RNA-seq data. Second, to address the imbalance-between-modalities problem, we propose a gradient modulation mechanism tailored to the Cox partial likelihood loss for survival prediction. The contributions of the modalities are dynamically monitored and adjusted during the training process, encouraging that both modalities are sufficiently trained. Evaluated on two TCGA(The Cancer Genome Atlas) datasets, our model achieves substantially improved survival prediction accuracy.
PHG-Net: Persistent Homology Guided Medical Image Classification
Nov 28, 2023



Abstract:Modern deep neural networks have achieved great successes in medical image analysis. However, the features captured by convolutional neural networks (CNNs) or Transformers tend to be optimized for pixel intensities and neglect key anatomical structures such as connected components and loops. In this paper, we propose a persistent homology guided approach (PHG-Net) that explores topological features of objects for medical image classification. For an input image, we first compute its cubical persistence diagram and extract topological features into a vector representation using a small neural network (called the PH module). The extracted topological features are then incorporated into the feature map generated by CNN or Transformer for feature fusion. The PH module is lightweight and capable of integrating topological features into any CNN or Transformer architectures in an end-to-end fashion. We evaluate our PHG-Net on three public datasets and demonstrate its considerable improvements on the target classification tasks over state-of-the-art methods.
Securer and Faster Privacy-Preserving Distributed Machine Learning
Nov 17, 2022Abstract:With the development of machine learning, it is difficult for a single server to process all the data. So machine learning tasks need to be spread across multiple servers, turning centralized machine learning into a distributed one. However, privacy remains an unsolved problem in distributed machine learning. Multi-key homomorphic encryption over torus (MKTFHE) is one of the suitable candidates to solve the problem. However, there may be security risks in the decryption of MKTFHE and the most recent result about MKFHE only supports the Boolean operation and linear operation. So, MKTFHE cannot compute the non-linear function like Sigmoid directly and it is still hard to perform common machine learning such as logistic regression and neural networks in high performance. This paper first introduces secret sharing to propose a new distributed decryption protocol for MKTFHE, then designs an MKTFHE-friendly activation function, and finally utilizes them to implement logistic regression and neural network training in MKTFHE. We prove the correctness and security of our decryption protocol and compare the efficiency and accuracy between using Taylor polynomials of Sigmoid and our proposed function as an activation function. The experiments show that the efficiency of our function is 10 times higher than using 7-order Taylor polynomials straightly and the accuracy of the training model is similar to that of using a high-order polynomial as an activation function scheme.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge