Ji Shi
CVeDRL: An Efficient Code Verifier via Difficulty-aware Reinforcement Learning
Jan 30, 2026Abstract:Code verifiers play a critical role in post-verification for LLM-based code generation, yet existing supervised fine-tuning methods suffer from data scarcity, high failure rates, and poor inference efficiency. While reinforcement learning (RL) offers a promising alternative by optimizing models through execution-driven rewards without labeled supervision, our preliminary results show that naive RL with only functionality rewards fails to generate effective unit tests for difficult branches and samples. We first theoretically analyze showing that branch coverage, sample difficulty, syntactic and functional correctness can be jointly modeled as RL rewards, where optimizing these signals can improve the reliability of unit-test-based verification. Guided by this analysis, we design syntax- and functionality-aware rewards and further propose branch- and sample-difficulty--aware RL using exponential reward shaping and static analysis metrics. With this formulation, CVeDRL achieves state-of-the-art performance with only 0.6B parameters, yielding up to 28.97% higher pass rate and 15.08% higher branch coverage than GPT-3.5, while delivering over $20\times$ faster inference than competitive baselines. Code is available at https://github.com/LIGHTCHASER1/CVeDRL.git
vLinear: A Powerful Linear Model for Multivariate Time Series Forecasting
Jan 20, 2026Abstract:In this paper, we present \textbf{vLinear}, an effective yet efficient \textbf{linear}-based multivariate time series forecaster featuring two components: the \textbf{v}ecTrans module and the WFMLoss objective. Many state-of-the-art forecasters rely on self-attention or its variants to capture multivariate correlations, typically incurring $\mathcal{O}(N^2)$ computational complexity with respect to the number of variates $N$. To address this, we propose vecTrans, a lightweight module that utilizes a learnable vector to model multivariate correlations, reducing the complexity to $\mathcal{O}(N)$. Notably, vecTrans can be seamlessly integrated into Transformer-based forecasters, delivering up to 5$\times$ inference speedups and consistent performance gains. Furthermore, we introduce WFMLoss (Weighted Flow Matching Loss) as the objective. In contrast to typical \textbf{velocity-oriented} flow matching objectives, we demonstrate that a \textbf{final-series-oriented} formulation yields significantly superior forecasting accuracy. WFMLoss also incorporates path- and horizon-weighted strategies to focus learning on more reliable paths and horizons. Empirically, vLinear achieves state-of-the-art performance across 22 benchmarks and 124 forecasting settings. Moreover, WFMLoss serves as an effective plug-and-play objective, consistently improving existing forecasters. The code is available at https://anonymous.4open.science/r/vLinear.
Staged Voxel-Level Deep Reinforcement Learning for 3D Medical Image Segmentation with Noisy Annotations
Jan 07, 2026Abstract:Deep learning has achieved significant advancements in medical image segmentation. Currently, obtaining accurate segmentation outcomes is critically reliant on large-scale datasets with high-quality annotations. However, noisy annotations are frequently encountered owing to the complex morphological structures of organs in medical images and variations among different annotators, which can substantially limit the efficacy of segmentation models. Motivated by the fact that medical imaging annotator can correct labeling errors during segmentation based on prior knowledge, we propose an end-to-end Staged Voxel-Level Deep Reinforcement Learning (SVL-DRL) framework for robust medical image segmentation under noisy annotations. This framework employs a dynamic iterative update strategy to automatically mitigate the impact of erroneous labels without requiring manual intervention. The key advancements of SVL-DRL over existing works include: i) formulating noisy annotations as a voxel-dependent problem and addressing it through a novel staged reinforcement learning framework which guarantees robust model convergence; ii) incorporating a voxel-level asynchronous advantage actor-critic (vA3C) module that conceptualizes each voxel as an autonomous agent, which allows each agent to dynamically refine its own state representation during training, thereby directly mitigating the influence of erroneous labels; iii) designing a novel action space for the agents, along with a composite reward function that strategically combines the Dice value and a spatial continuity metric to significantly boost segmentation accuracy while maintain semantic integrity. Experiments on three public medical image datasets demonstrates State-of-The-Art (SoTA) performance under various experimental settings, with an average improvement of over 3\% in both Dice and IoU scores.
REACT-LLM: A Benchmark for Evaluating LLM Integration with Causal Features in Clinical Prognostic Tasks
Nov 13, 2025Abstract:Large Language Models (LLMs) and causal learning each hold strong potential for clinical decision making (CDM). However, their synergy remains poorly understood, largely due to the lack of systematic benchmarks evaluating their integration in clinical risk prediction. In real-world healthcare, identifying features with causal influence on outcomes is crucial for actionable and trustworthy predictions. While recent work highlights LLMs' emerging causal reasoning abilities, there lacks comprehensive benchmarks to assess their causal learning and performance informed by causal features in clinical risk prediction. To address this, we introduce REACT-LLM, a benchmark designed to evaluate whether combining LLMs with causal features can enhance clinical prognostic performance and potentially outperform traditional machine learning (ML) methods. Unlike existing LLM-clinical benchmarks that often focus on a limited set of outcomes, REACT-LLM evaluates 7 clinical outcomes across 2 real-world datasets, comparing 15 prominent LLMs, 6 traditional ML models, and 3 causal discovery (CD) algorithms. Our findings indicate that while LLMs perform reasonably in clinical prognostics, they have not yet outperformed traditional ML models. Integrating causal features derived from CD algorithms into LLMs offers limited performance gains, primarily due to the strict assumptions of many CD methods, which are often violated in complex clinical data. While the direct integration yields limited improvement, our benchmark reveals a more promising synergy.
Fast $k$-means clustering in Riemannian manifolds via Fréchet maps: Applications to large-dimensional SPD matrices
Nov 12, 2025Abstract:We introduce a novel, efficient framework for clustering data on high-dimensional, non-Euclidean manifolds that overcomes the computational challenges associated with standard intrinsic methods. The key innovation is the use of the $p$-Fréchet map $F^p : \mathcal{M} \to \mathbb{R}^\ell$ -- defined on a generic metric space $\mathcal{M}$ -- which embeds the manifold data into a lower-dimensional Euclidean space $\mathbb{R}^\ell$ using a set of reference points $\{r_i\}_{i=1}^\ell$, $r_i \in \mathcal{M}$. Once embedded, we can efficiently and accurately apply standard Euclidean clustering techniques such as k-means. We rigorously analyze the mathematical properties of $F^p$ in the Euclidean space and the challenging manifold of $n \times n$ symmetric positive definite matrices $\mathit{SPD}(n)$. Extensive numerical experiments using synthetic and real $\mathit{SPD}(n)$ data demonstrate significant performance gains: our method reduces runtime by up to two orders of magnitude compared to intrinsic manifold-based approaches, all while maintaining high clustering accuracy, including scenarios where existing alternative methods struggle or fail.
OLinear: A Linear Model for Time Series Forecasting in Orthogonally Transformed Domain
May 14, 2025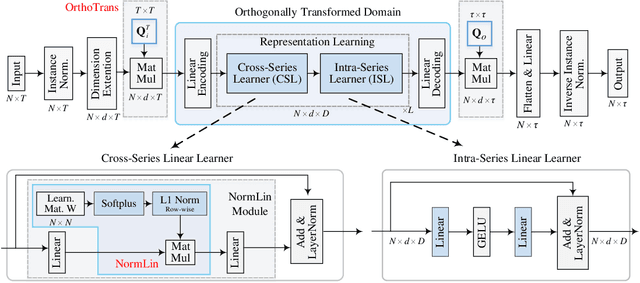
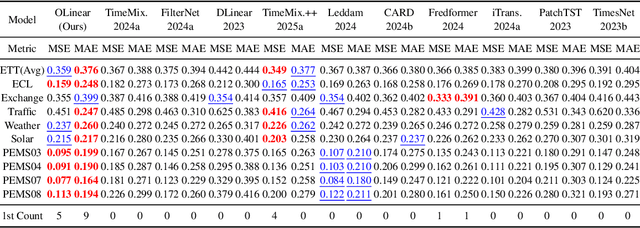
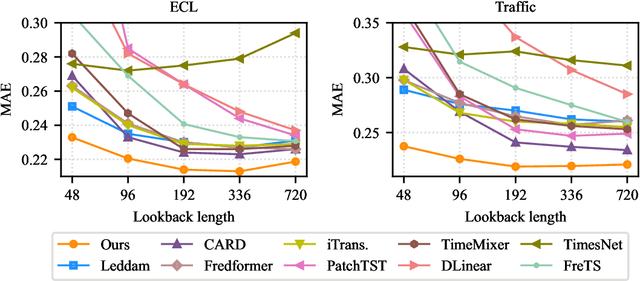
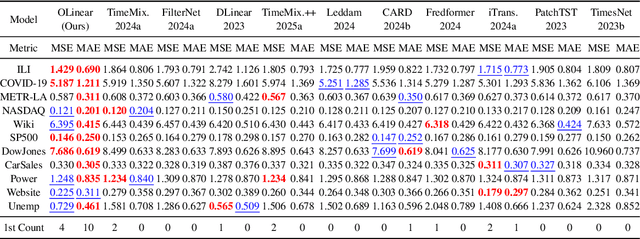
Abstract:This paper presents $\mathbf{OLinear}$, a $\mathbf{linear}$-based multivariate time series forecasting model that operates in an $\mathbf{o}$rthogonally transformed domain. Recent forecasting models typically adopt the temporal forecast (TF) paradigm, which directly encode and decode time series in the time domain. However, the entangled step-wise dependencies in series data can hinder the performance of TF. To address this, some forecasters conduct encoding and decoding in the transformed domain using fixed, dataset-independent bases (e.g., sine and cosine signals in the Fourier transform). In contrast, we utilize $\mathbf{OrthoTrans}$, a data-adaptive transformation based on an orthogonal matrix that diagonalizes the series' temporal Pearson correlation matrix. This approach enables more effective encoding and decoding in the decorrelated feature domain and can serve as a plug-in module to enhance existing forecasters. To enhance the representation learning for multivariate time series, we introduce a customized linear layer, $\mathbf{NormLin}$, which employs a normalized weight matrix to capture multivariate dependencies. Empirically, the NormLin module shows a surprising performance advantage over multi-head self-attention, while requiring nearly half the FLOPs. Extensive experiments on 24 benchmarks and 140 forecasting tasks demonstrate that OLinear consistently achieves state-of-the-art performance with high efficiency. Notably, as a plug-in replacement for self-attention, the NormLin module consistently enhances Transformer-based forecasters. The code and datasets are available at https://anonymous.4open.science/r/OLinear
JTreeformer: Graph-Transformer via Latent-Diffusion Model for Molecular Generation
Apr 29, 2025Abstract:The discovery of new molecules based on the original chemical molecule distributions is of great importance in medicine. The graph transformer, with its advantages of high performance and scalability compared to traditional graph networks, has been widely explored in recent research for applications of graph structures. However, current transformer-based graph decoders struggle to effectively utilize graph information, which limits their capacity to leverage only sequences of nodes rather than the complex topological structures of molecule graphs. This paper focuses on building a graph transformer-based framework for molecular generation, which we call \textbf{JTreeformer} as it transforms graph generation into junction tree generation. It combines GCN parallel with multi-head attention as the encoder. It integrates a directed acyclic GCN into a graph-based Transformer to serve as a decoder, which can iteratively synthesize the entire molecule by leveraging information from the partially constructed molecular structure at each step. In addition, a diffusion model is inserted in the latent space generated by the encoder, to enhance the efficiency and effectiveness of sampling further. The empirical results demonstrate that our novel framework outperforms existing molecule generation methods, thus offering a promising tool to advance drug discovery (https://anonymous.4open.science/r/JTreeformer-C74C).
SFi-Former: Sparse Flow Induced Attention for Graph Transformer
Apr 29, 2025Abstract:Graph Transformers (GTs) have demonstrated superior performance compared to traditional message-passing graph neural networks in many studies, especially in processing graph data with long-range dependencies. However, GTs tend to suffer from weak inductive bias, overfitting and over-globalizing problems due to the dense attention. In this paper, we introduce SFi-attention, a novel attention mechanism designed to learn sparse pattern by minimizing an energy function based on network flows with l1-norm regularization, to relieve those issues caused by dense attention. Furthermore, SFi-Former is accordingly devised which can leverage the sparse attention pattern of SFi-attention to generate sparse network flows beyond adjacency matrix of graph data. Specifically, SFi-Former aggregates features selectively from other nodes through flexible adaptation of the sparse attention, leading to a more robust model. We validate our SFi-Former on various graph datasets, especially those graph data exhibiting long-range dependencies. Experimental results show that our SFi-Former obtains competitive performance on GNN Benchmark datasets and SOTA performance on LongRange Graph Benchmark (LRGB) datasets. Additionally, our model gives rise to smaller generalization gaps, which indicates that it is less prone to over-fitting. Click here for codes.
A Smooth Analytical Formulation of Collision Detection and Rigid Body Dynamics With Contact
Mar 14, 2025Abstract:Generating intelligent robot behavior in contact-rich settings is a research problem where zeroth-order methods currently prevail. A major contributor to the success of such methods is their robustness in the face of non-smooth and discontinuous optimization landscapes that are characteristic of contact interactions, yet zeroth-order methods remain computationally inefficient. It is therefore desirable to develop methods for perception, planning and control in contact-rich settings that can achieve further efficiency by making use of first and second order information (i.e., gradients and Hessians). To facilitate this, we present a joint formulation of collision detection and contact modelling which, compared to existing differentiable simulation approaches, provides the following benefits: i) it results in forward and inverse dynamics that are entirely analytical (i.e. do not require solving optimization or root-finding problems with iterative methods) and smooth (i.e. twice differentiable), ii) it supports arbitrary collision geometries without needing a convex decomposition, and iii) its runtime is independent of the number of contacts. Through simulation experiments, we demonstrate the validity of the proposed formulation as a "physics for inference" that can facilitate future development of efficient methods to generate intelligent contact-rich behavior.
GIGP: A Global Information Interacting and Geometric Priors Focusing Framework for Semi-supervised Medical Image Segmentation
Mar 12, 2025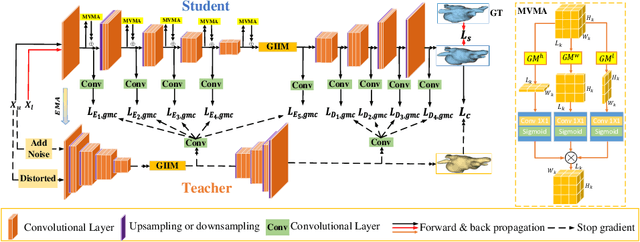
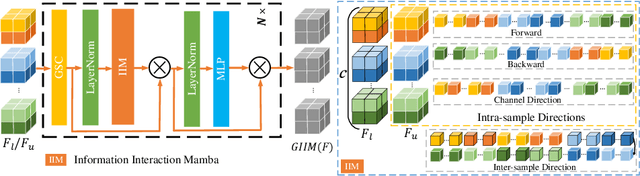
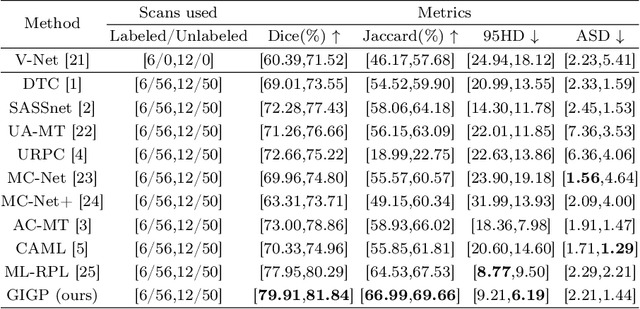
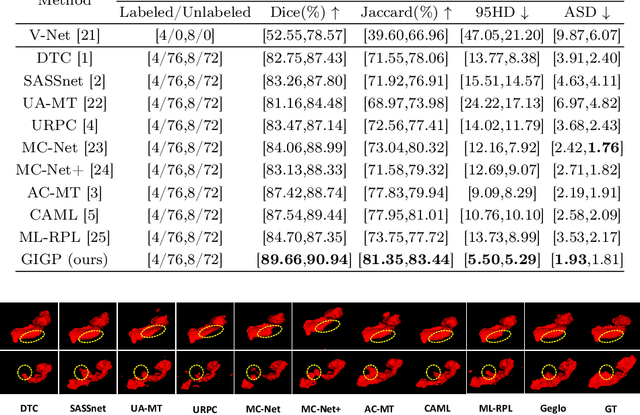
Abstract:Semi-supervised learning enhances medical image segmentation by leveraging unlabeled data, reducing reliance on extensive labeled datasets. On the one hand, the distribution discrepancy between limited labeled data and abundant unlabeled data can hinder model generalization. Most existing methods rely on local similarity matching, which may introduce bias. In contrast, Mamba effectively models global context with linear complexity, learning more comprehensive data representations. On the other hand, medical images usually exhibit consistent anatomical structures defined by geometric features. Most existing methods fail to fully utilize global geometric priors, such as volumes, moments etc. In this work, we introduce a global information interaction and geometric priors focus framework (GIGP). Firstly, we present a Global Information Interaction Mamba module to reduce distribution discrepancy between labeled and unlabeled data. Secondly, we propose a Geometric Moment Attention Mechanism to extract richer global geometric features. Finally, we propose Global Geometric Perturbation Consistency to simulate organ dynamics and geometric variations, enhancing the ability of the model to learn generalized features. The superior performance on the NIH Pancreas and Left Atrium datasets demonstrates the effectiveness of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge