Lianyuan Yu
GIGP: A Global Information Interacting and Geometric Priors Focusing Framework for Semi-supervised Medical Image Segmentation
Mar 12, 2025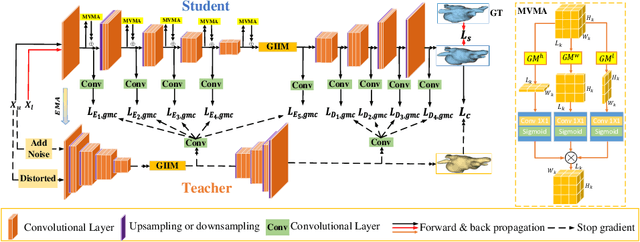
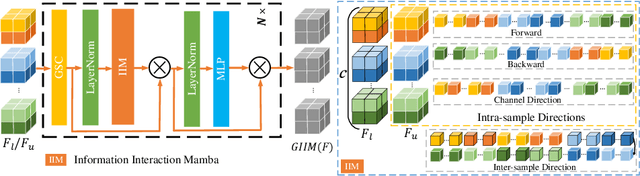
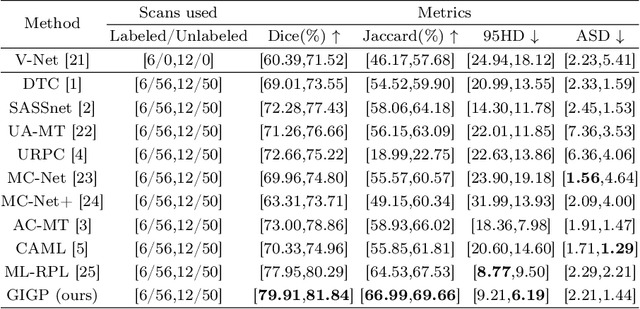
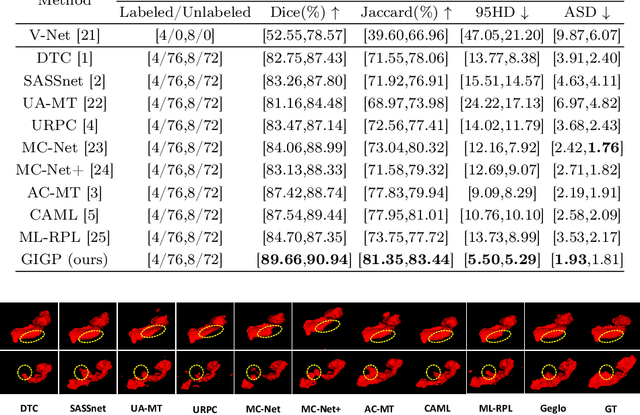
Abstract:Semi-supervised learning enhances medical image segmentation by leveraging unlabeled data, reducing reliance on extensive labeled datasets. On the one hand, the distribution discrepancy between limited labeled data and abundant unlabeled data can hinder model generalization. Most existing methods rely on local similarity matching, which may introduce bias. In contrast, Mamba effectively models global context with linear complexity, learning more comprehensive data representations. On the other hand, medical images usually exhibit consistent anatomical structures defined by geometric features. Most existing methods fail to fully utilize global geometric priors, such as volumes, moments etc. In this work, we introduce a global information interaction and geometric priors focus framework (GIGP). Firstly, we present a Global Information Interaction Mamba module to reduce distribution discrepancy between labeled and unlabeled data. Secondly, we propose a Geometric Moment Attention Mechanism to extract richer global geometric features. Finally, we propose Global Geometric Perturbation Consistency to simulate organ dynamics and geometric variations, enhancing the ability of the model to learn generalized features. The superior performance on the NIH Pancreas and Left Atrium datasets demonstrates the effectiveness of our approach.
Diff-CL: A Novel Cross Pseudo-Supervision Method for Semi-supervised Medical Image Segmentation
Mar 12, 2025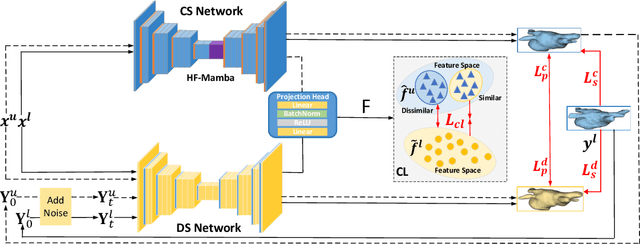
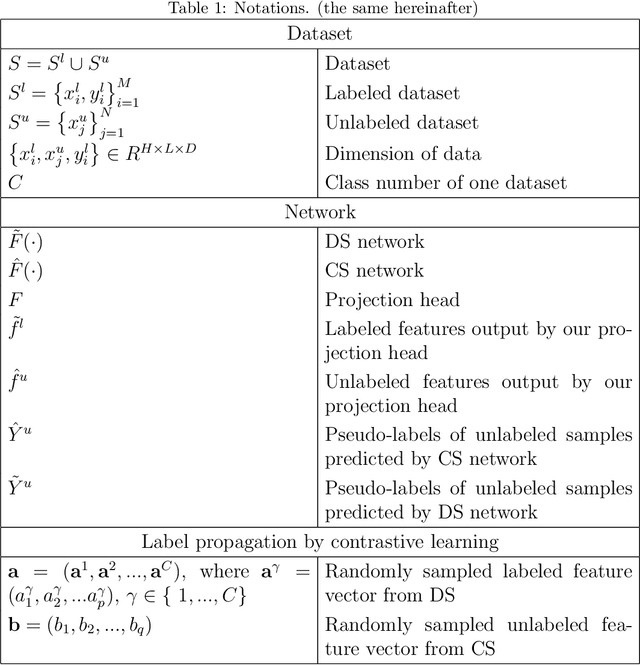
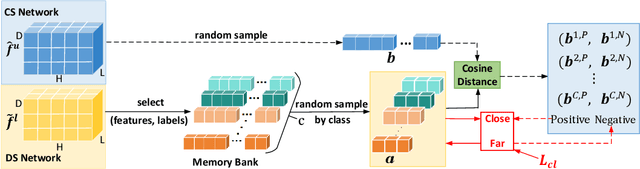
Abstract:Semi-supervised learning utilizes insights from unlabeled data to improve model generalization, thereby reducing reliance on large labeled datasets. Most existing studies focus on limited samples and fail to capture the overall data distribution. We contend that combining distributional information with detailed information is crucial for achieving more robust and accurate segmentation results. On the one hand, with its robust generative capabilities, diffusion models (DM) learn data distribution effectively. However, it struggles with fine detail capture, leading to generated images with misleading details. Combining DM with convolutional neural networks (CNNs) enables the former to learn data distribution while the latter corrects fine details. While capturing complete high-frequency details by CNNs requires substantial computational resources and is susceptible to local noise. On the other hand, given that both labeled and unlabeled data come from the same distribution, we believe that regions in unlabeled data similar to overall class semantics to labeled data are likely to belong to the same class, while regions with minimal similarity are less likely to. This work introduces a semi-supervised medical image segmentation framework from the distribution perspective (Diff-CL). Firstly, we propose a cross-pseudo-supervision learning mechanism between diffusion and convolution segmentation networks. Secondly, we design a high-frequency mamba module to capture boundary and detail information globally. Finally, we apply contrastive learning for label propagation from labeled to unlabeled data. Our method achieves state-of-the-art (SOTA) performance across three datasets, including left atrium, brain tumor, and NIH pancreas datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge