Chaoli Wang
An Evaluation-Centric Paradigm for Scientific Visualization Agents
Sep 18, 2025
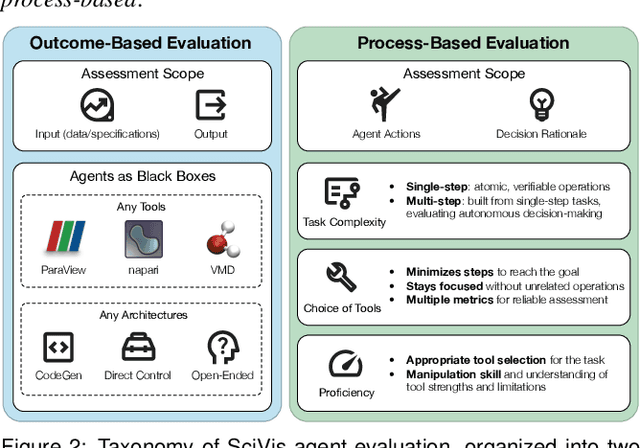
Abstract:Recent advances in multi-modal large language models (MLLMs) have enabled increasingly sophisticated autonomous visualization agents capable of translating user intentions into data visualizations. However, measuring progress and comparing different agents remains challenging, particularly in scientific visualization (SciVis), due to the absence of comprehensive, large-scale benchmarks for evaluating real-world capabilities. This position paper examines the various types of evaluation required for SciVis agents, outlines the associated challenges, provides a simple proof-of-concept evaluation example, and discusses how evaluation benchmarks can facilitate agent self-improvement. We advocate for a broader collaboration to develop a SciVis agentic evaluation benchmark that would not only assess existing capabilities but also drive innovation and stimulate future development in the field.
TexGS-VolVis: Expressive Scene Editing for Volume Visualization via Textured Gaussian Splatting
Jul 18, 2025Abstract:Advancements in volume visualization (VolVis) focus on extracting insights from 3D volumetric data by generating visually compelling renderings that reveal complex internal structures. Existing VolVis approaches have explored non-photorealistic rendering techniques to enhance the clarity, expressiveness, and informativeness of visual communication. While effective, these methods often rely on complex predefined rules and are limited to transferring a single style, restricting their flexibility. To overcome these limitations, we advocate the representation of VolVis scenes using differentiable Gaussian primitives combined with pretrained large models to enable arbitrary style transfer and real-time rendering. However, conventional 3D Gaussian primitives tightly couple geometry and appearance, leading to suboptimal stylization results. To address this, we introduce TexGS-VolVis, a textured Gaussian splatting framework for VolVis. TexGS-VolVis employs 2D Gaussian primitives, extending each Gaussian with additional texture and shading attributes, resulting in higher-quality, geometry-consistent stylization and enhanced lighting control during inference. Despite these improvements, achieving flexible and controllable scene editing remains challenging. To further enhance stylization, we develop image- and text-driven non-photorealistic scene editing tailored for TexGS-VolVis and 2D-lift-3D segmentation to enable partial editing with fine-grained control. We evaluate TexGS-VolVis both qualitatively and quantitatively across various volume rendering scenes, demonstrating its superiority over existing methods in terms of efficiency, visual quality, and editing flexibility.
MC-INR: Efficient Encoding of Multivariate Scientific Simulation Data using Meta-Learning and Clustered Implicit Neural Representations
Jul 03, 2025Abstract:Implicit Neural Representations (INRs) are widely used to encode data as continuous functions, enabling the visualization of large-scale multivariate scientific simulation data with reduced memory usage. However, existing INR-based methods face three main limitations: (1) inflexible representation of complex structures, (2) primarily focusing on single-variable data, and (3) dependence on structured grids. Thus, their performance degrades when applied to complex real-world datasets. To address these limitations, we propose a novel neural network-based framework, MC-INR, which handles multivariate data on unstructured grids. It combines meta-learning and clustering to enable flexible encoding of complex structures. To further improve performance, we introduce a residual-based dynamic re-clustering mechanism that adaptively partitions clusters based on local error. We also propose a branched layer to leverage multivariate data through independent branches simultaneously. Experimental results demonstrate that MC-INR outperforms existing methods on scientific data encoding tasks.
iVR-GS: Inverse Volume Rendering for Explorable Visualization via Editable 3D Gaussian Splatting
Apr 24, 2025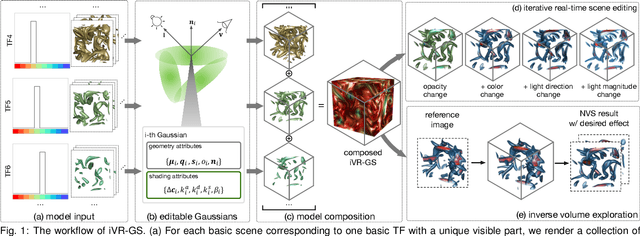

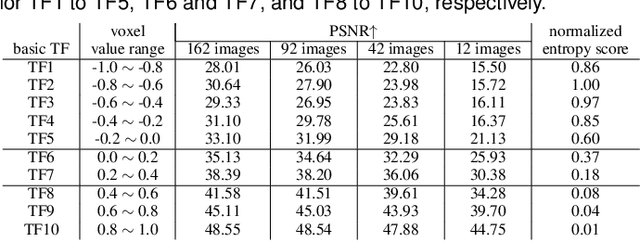
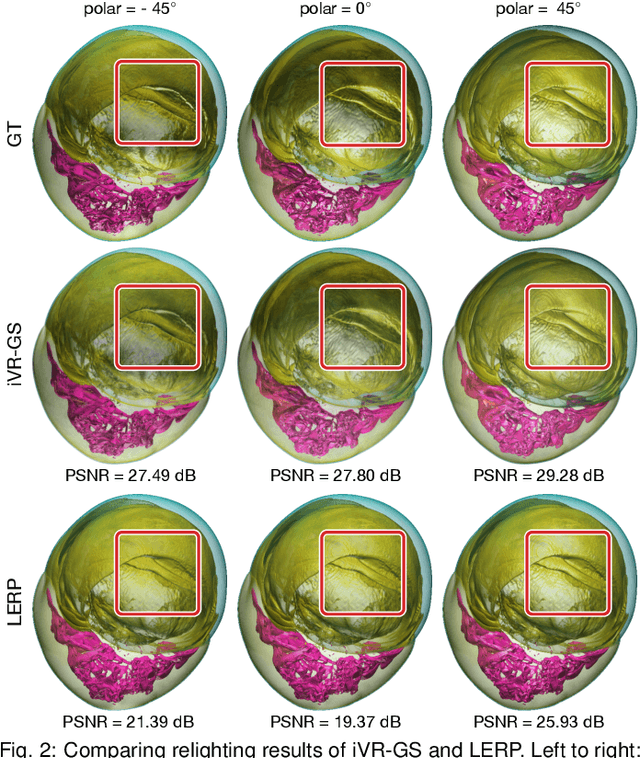
Abstract:In volume visualization, users can interactively explore the three-dimensional data by specifying color and opacity mappings in the transfer function (TF) or adjusting lighting parameters, facilitating meaningful interpretation of the underlying structure. However, rendering large-scale volumes demands powerful GPUs and high-speed memory access for real-time performance. While existing novel view synthesis (NVS) methods offer faster rendering speeds with lower hardware requirements, the visible parts of a reconstructed scene are fixed and constrained by preset TF settings, significantly limiting user exploration. This paper introduces inverse volume rendering via Gaussian splatting (iVR-GS), an innovative NVS method that reduces the rendering cost while enabling scene editing for interactive volume exploration. Specifically, we compose multiple iVR-GS models associated with basic TFs covering disjoint visible parts to make the entire volumetric scene visible. Each basic model contains a collection of 3D editable Gaussians, where each Gaussian is a 3D spatial point that supports real-time scene rendering and editing. We demonstrate the superior reconstruction quality and composability of iVR-GS against other NVS solutions (Plenoxels, CCNeRF, and base 3DGS) on various volume datasets. The code is available at https://github.com/TouKaienn/iVR-GS.
AI-Powered Automated Model Construction for Patient-Specific CFD Simulations of Aortic Flows
Mar 16, 2025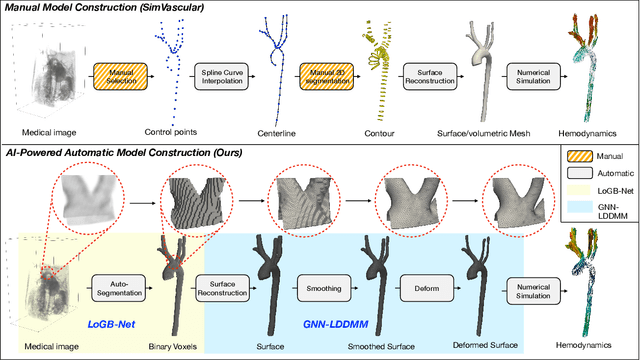
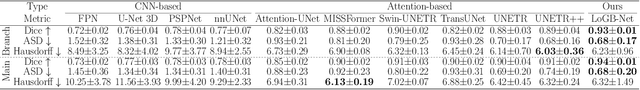
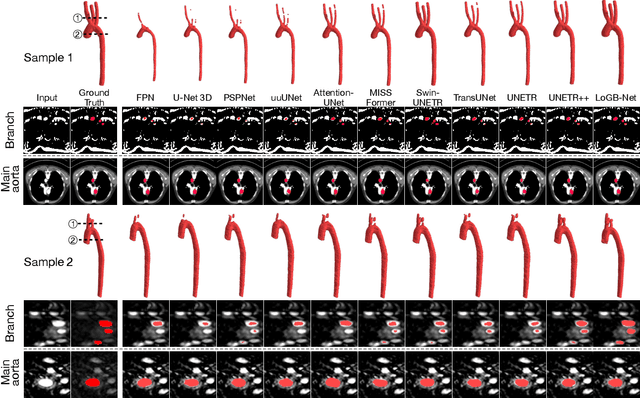
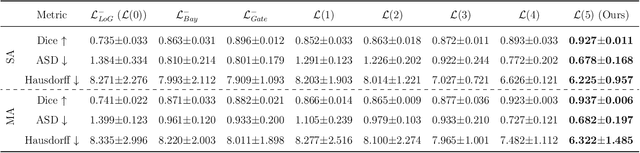
Abstract:Image-based modeling is essential for understanding cardiovascular hemodynamics and advancing the diagnosis and treatment of cardiovascular diseases. Constructing patient-specific vascular models remains labor-intensive, error-prone, and time-consuming, limiting their clinical applications. This study introduces a deep-learning framework that automates the creation of simulation-ready vascular models from medical images. The framework integrates a segmentation module for accurate voxel-based vessel delineation with a surface deformation module that performs anatomically consistent and unsupervised surface refinements guided by medical image data. By unifying voxel segmentation and surface deformation into a single cohesive pipeline, the framework addresses key limitations of existing methods, enhancing geometric accuracy and computational efficiency. Evaluated on publicly available datasets, the proposed approach demonstrates state-of-the-art performance in segmentation and mesh quality while significantly reducing manual effort and processing time. This work advances the scalability and reliability of image-based computational modeling, facilitating broader applications in clinical and research settings.
Hierarchical LoG Bayesian Neural Network for Enhanced Aorta Segmentation
Jan 18, 2025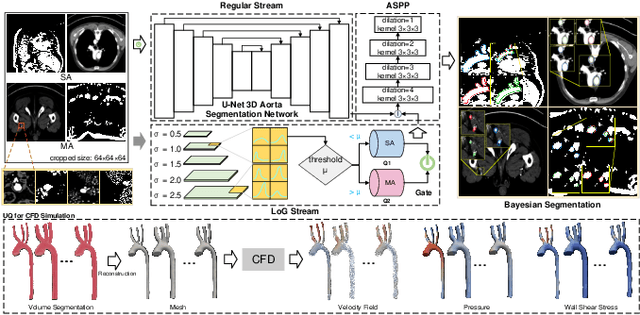

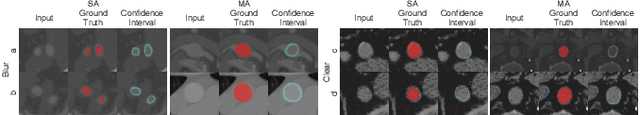
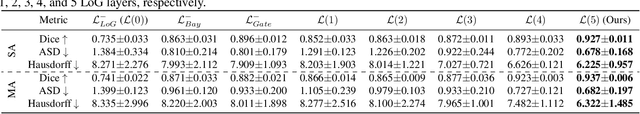
Abstract:Accurate segmentation of the aorta and its associated arch branches is crucial for diagnosing aortic diseases. While deep learning techniques have significantly improved aorta segmentation, they remain challenging due to the intricate multiscale structure and the complexity of the surrounding tissues. This paper presents a novel approach for enhancing aorta segmentation using a Bayesian neural network-based hierarchical Laplacian of Gaussian (LoG) model. Our model consists of a 3D U-Net stream and a hierarchical LoG stream: the former provides an initial aorta segmentation, and the latter enhances blood vessel detection across varying scales by learning suitable LoG kernels, enabling self-adaptive handling of different parts of the aorta vessels with significant scale differences. We employ a Bayesian method to parameterize the LoG stream and provide confidence intervals for the segmentation results, ensuring robustness and reliability of the prediction for vascular medical image analysts. Experimental results show that our model can accurately segment main and supra-aortic vessels, yielding at least a 3% gain in the Dice coefficient over state-of-the-art methods across multiple volumes drawn from two aorta datasets, and can provide reliable confidence intervals for different parts of the aorta. The code is available at https://github.com/adlsn/LoGBNet.
SurfPatch: Enabling Patch Matching for Exploratory Stream Surface Visualization
Jan 01, 2025



Abstract:Unlike their line-based counterparts, surface-based techniques have yet to be thoroughly investigated in flow visualization due to their significant placement, speed, perception, and evaluation challenges. This paper presents SurfPatch, a novel framework supporting exploratory stream surface visualization. To begin with, we translate the issue of surface placement to surface selection and trace a large number of stream surfaces from a given flow field dataset. Then, we introduce a three-stage process: vertex-level classification, patch-level matching, and surface-level clustering that hierarchically builds the connection between vertices and patches and between patches and surfaces. This bottom-up approach enables fine-grained, multiscale patch-level matching, sharply contrasts surface-level matching offered by existing works, and provides previously unavailable flexibility during querying. We design an intuitive visual interface for users to conveniently visualize and analyze the underlying collection of stream surfaces in an exploratory manner. SurfPatch is not limited to stream surfaces traced from steady flow datasets. We demonstrate its effectiveness through experiments on stream surfaces produced from steady and unsteady flows as well as isosurfaces extracted from scalar fields. The code is available at https://github.com/adlsn/SurfPatch.
Sli2Vol+: Segmenting 3D Medical Images Based on an Object Estimation Guided Correspondence Flow Network
Nov 21, 2024



Abstract:Deep learning (DL) methods have shown remarkable successes in medical image segmentation, often using large amounts of annotated data for model training. However, acquiring a large number of diverse labeled 3D medical image datasets is highly difficult and expensive. Recently, mask propagation DL methods were developed to reduce the annotation burden on 3D medical images. For example, Sli2Vol~\cite{yeung2021sli2vol} proposed a self-supervised framework (SSF) to learn correspondences by matching neighboring slices via slice reconstruction in the training stage; the learned correspondences were then used to propagate a labeled slice to other slices in the test stage. But, these methods are still prone to error accumulation due to the inter-slice propagation of reconstruction errors. Also, they do not handle discontinuities well, which can occur between consecutive slices in 3D images, as they emphasize exploiting object continuity. To address these challenges, in this work, we propose a new SSF, called \proposed, {for segmenting any anatomical structures in 3D medical images using only a single annotated slice per training and testing volume.} Specifically, in the training stage, we first propagate an annotated 2D slice of a training volume to the other slices, generating pseudo-labels (PLs). Then, we develop a novel Object Estimation Guided Correspondence Flow Network to learn reliable correspondences between consecutive slices and corresponding PLs in a self-supervised manner. In the test stage, such correspondences are utilized to propagate a single annotated slice to the other slices of a test volume. We demonstrate the effectiveness of our method on various medical image segmentation tasks with different datasets, showing better generalizability across different organs, modalities, and modals. Code is available at \url{https://github.com/adlsn/Sli2Volplus}
StyleRF-VolVis: Style Transfer of Neural Radiance Fields for Expressive Volume Visualization
Jul 31, 2024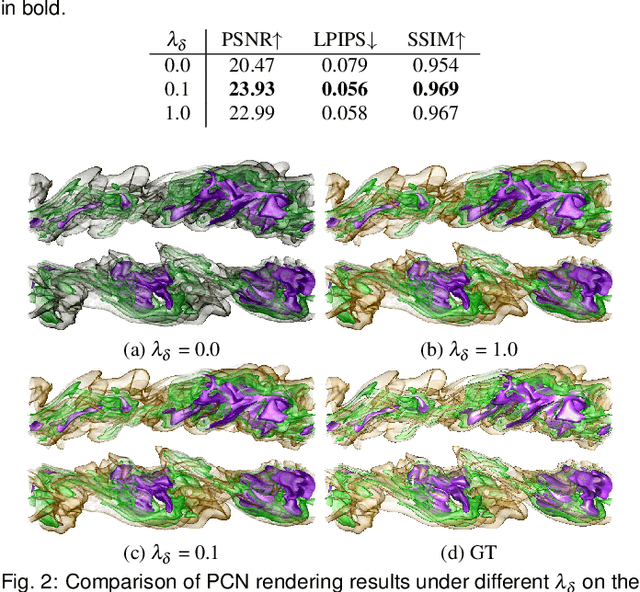
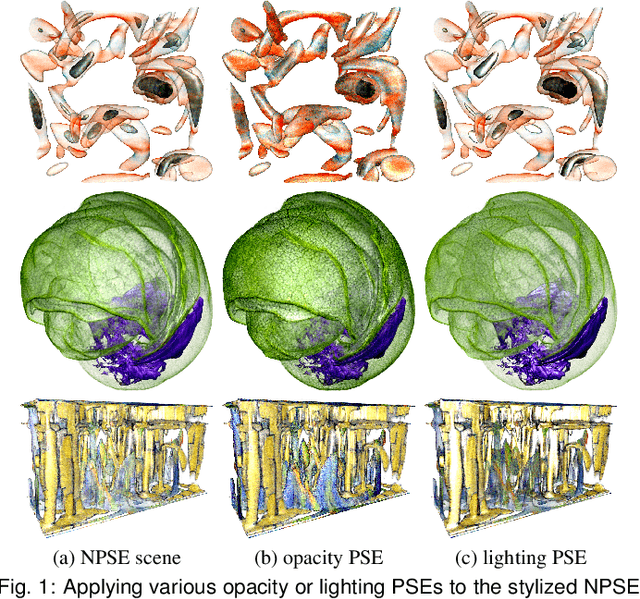
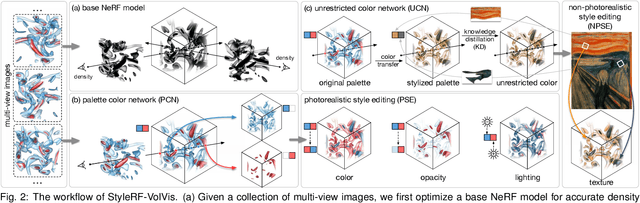

Abstract:In volume visualization, visualization synthesis has attracted much attention due to its ability to generate novel visualizations without following the conventional rendering pipeline. However, existing solutions based on generative adversarial networks often require many training images and take significant training time. Still, issues such as low quality, consistency, and flexibility persist. This paper introduces StyleRF-VolVis, an innovative style transfer framework for expressive volume visualization (VolVis) via neural radiance field (NeRF). The expressiveness of StyleRF-VolVis is upheld by its ability to accurately separate the underlying scene geometry (i.e., content) and color appearance (i.e., style), conveniently modify color, opacity, and lighting of the original rendering while maintaining visual content consistency across the views, and effectively transfer arbitrary styles from reference images to the reconstructed 3D scene. To achieve these, we design a base NeRF model for scene geometry extraction, a palette color network to classify regions of the radiance field for photorealistic editing, and an unrestricted color network to lift the color palette constraint via knowledge distillation for non-photorealistic editing. We demonstrate the superior quality, consistency, and flexibility of StyleRF-VolVis by experimenting with various volume rendering scenes and reference images and comparing StyleRF-VolVis against other image-based (AdaIN), video-based (ReReVST), and NeRF-based (ARF and SNeRF) style rendering solutions.
A Comparative Study of Neural Surface Reconstruction for Scientific Visualization
Jul 30, 2024Abstract:This comparative study evaluates various neural surface reconstruction methods, particularly focusing on their implications for scientific visualization through reconstructing 3D surfaces via multi-view rendering images. We categorize ten methods into neural radiance fields and neural implicit surfaces, uncovering the benefits of leveraging distance functions (i.e., SDFs and UDFs) to enhance the accuracy and smoothness of the reconstructed surfaces. Our findings highlight the efficiency and quality of NeuS2 for reconstructing closed surfaces and identify NeUDF as a promising candidate for reconstructing open surfaces despite some limitations. By sharing our benchmark dataset, we invite researchers to test the performance of their methods, contributing to the advancement of surface reconstruction solutions for scientific visualization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge