Delin An
AI-Powered Automated Model Construction for Patient-Specific CFD Simulations of Aortic Flows
Mar 16, 2025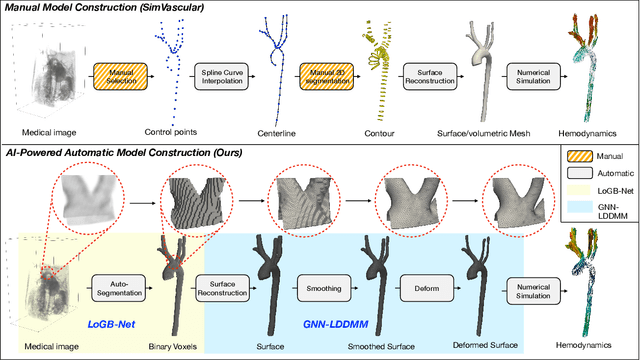
Abstract:Image-based modeling is essential for understanding cardiovascular hemodynamics and advancing the diagnosis and treatment of cardiovascular diseases. Constructing patient-specific vascular models remains labor-intensive, error-prone, and time-consuming, limiting their clinical applications. This study introduces a deep-learning framework that automates the creation of simulation-ready vascular models from medical images. The framework integrates a segmentation module for accurate voxel-based vessel delineation with a surface deformation module that performs anatomically consistent and unsupervised surface refinements guided by medical image data. By unifying voxel segmentation and surface deformation into a single cohesive pipeline, the framework addresses key limitations of existing methods, enhancing geometric accuracy and computational efficiency. Evaluated on publicly available datasets, the proposed approach demonstrates state-of-the-art performance in segmentation and mesh quality while significantly reducing manual effort and processing time. This work advances the scalability and reliability of image-based computational modeling, facilitating broader applications in clinical and research settings.
Hierarchical LoG Bayesian Neural Network for Enhanced Aorta Segmentation
Jan 18, 2025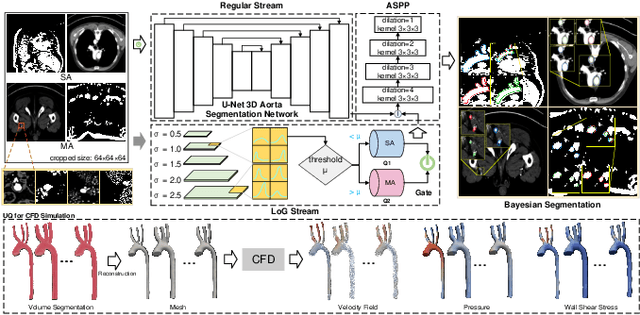

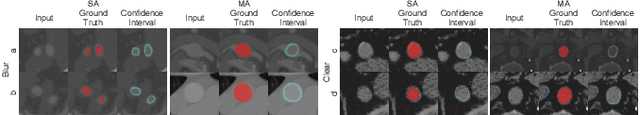
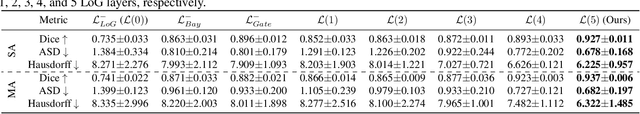
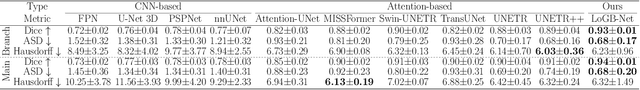
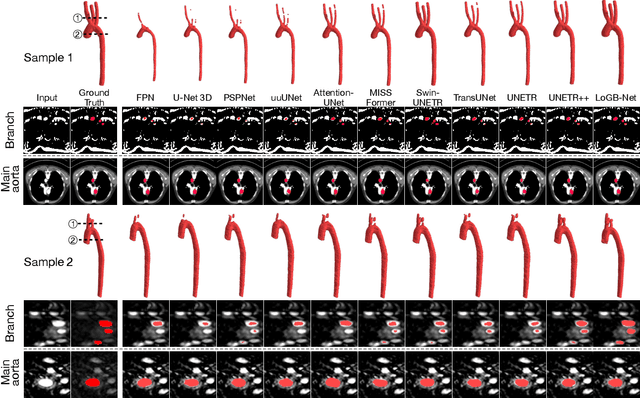
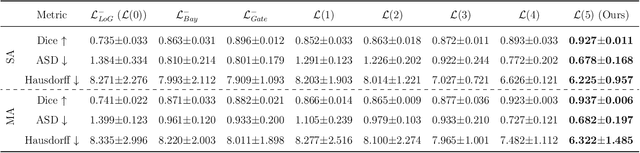
Abstract:Accurate segmentation of the aorta and its associated arch branches is crucial for diagnosing aortic diseases. While deep learning techniques have significantly improved aorta segmentation, they remain challenging due to the intricate multiscale structure and the complexity of the surrounding tissues. This paper presents a novel approach for enhancing aorta segmentation using a Bayesian neural network-based hierarchical Laplacian of Gaussian (LoG) model. Our model consists of a 3D U-Net stream and a hierarchical LoG stream: the former provides an initial aorta segmentation, and the latter enhances blood vessel detection across varying scales by learning suitable LoG kernels, enabling self-adaptive handling of different parts of the aorta vessels with significant scale differences. We employ a Bayesian method to parameterize the LoG stream and provide confidence intervals for the segmentation results, ensuring robustness and reliability of the prediction for vascular medical image analysts. Experimental results show that our model can accurately segment main and supra-aortic vessels, yielding at least a 3% gain in the Dice coefficient over state-of-the-art methods across multiple volumes drawn from two aorta datasets, and can provide reliable confidence intervals for different parts of the aorta. The code is available at https://github.com/adlsn/LoGBNet.
SurfPatch: Enabling Patch Matching for Exploratory Stream Surface Visualization
Jan 01, 2025



Abstract:Unlike their line-based counterparts, surface-based techniques have yet to be thoroughly investigated in flow visualization due to their significant placement, speed, perception, and evaluation challenges. This paper presents SurfPatch, a novel framework supporting exploratory stream surface visualization. To begin with, we translate the issue of surface placement to surface selection and trace a large number of stream surfaces from a given flow field dataset. Then, we introduce a three-stage process: vertex-level classification, patch-level matching, and surface-level clustering that hierarchically builds the connection between vertices and patches and between patches and surfaces. This bottom-up approach enables fine-grained, multiscale patch-level matching, sharply contrasts surface-level matching offered by existing works, and provides previously unavailable flexibility during querying. We design an intuitive visual interface for users to conveniently visualize and analyze the underlying collection of stream surfaces in an exploratory manner. SurfPatch is not limited to stream surfaces traced from steady flow datasets. We demonstrate its effectiveness through experiments on stream surfaces produced from steady and unsteady flows as well as isosurfaces extracted from scalar fields. The code is available at https://github.com/adlsn/SurfPatch.
Sli2Vol+: Segmenting 3D Medical Images Based on an Object Estimation Guided Correspondence Flow Network
Nov 21, 2024



Abstract:Deep learning (DL) methods have shown remarkable successes in medical image segmentation, often using large amounts of annotated data for model training. However, acquiring a large number of diverse labeled 3D medical image datasets is highly difficult and expensive. Recently, mask propagation DL methods were developed to reduce the annotation burden on 3D medical images. For example, Sli2Vol~\cite{yeung2021sli2vol} proposed a self-supervised framework (SSF) to learn correspondences by matching neighboring slices via slice reconstruction in the training stage; the learned correspondences were then used to propagate a labeled slice to other slices in the test stage. But, these methods are still prone to error accumulation due to the inter-slice propagation of reconstruction errors. Also, they do not handle discontinuities well, which can occur between consecutive slices in 3D images, as they emphasize exploiting object continuity. To address these challenges, in this work, we propose a new SSF, called \proposed, {for segmenting any anatomical structures in 3D medical images using only a single annotated slice per training and testing volume.} Specifically, in the training stage, we first propagate an annotated 2D slice of a training volume to the other slices, generating pseudo-labels (PLs). Then, we develop a novel Object Estimation Guided Correspondence Flow Network to learn reliable correspondences between consecutive slices and corresponding PLs in a self-supervised manner. In the test stage, such correspondences are utilized to propagate a single annotated slice to the other slices of a test volume. We demonstrate the effectiveness of our method on various medical image segmentation tasks with different datasets, showing better generalizability across different organs, modalities, and modals. Code is available at \url{https://github.com/adlsn/Sli2Volplus}
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge