Milan Sonka
Sli2Vol+: Segmenting 3D Medical Images Based on an Object Estimation Guided Correspondence Flow Network
Nov 21, 2024



Abstract:Deep learning (DL) methods have shown remarkable successes in medical image segmentation, often using large amounts of annotated data for model training. However, acquiring a large number of diverse labeled 3D medical image datasets is highly difficult and expensive. Recently, mask propagation DL methods were developed to reduce the annotation burden on 3D medical images. For example, Sli2Vol~\cite{yeung2021sli2vol} proposed a self-supervised framework (SSF) to learn correspondences by matching neighboring slices via slice reconstruction in the training stage; the learned correspondences were then used to propagate a labeled slice to other slices in the test stage. But, these methods are still prone to error accumulation due to the inter-slice propagation of reconstruction errors. Also, they do not handle discontinuities well, which can occur between consecutive slices in 3D images, as they emphasize exploiting object continuity. To address these challenges, in this work, we propose a new SSF, called \proposed, {for segmenting any anatomical structures in 3D medical images using only a single annotated slice per training and testing volume.} Specifically, in the training stage, we first propagate an annotated 2D slice of a training volume to the other slices, generating pseudo-labels (PLs). Then, we develop a novel Object Estimation Guided Correspondence Flow Network to learn reliable correspondences between consecutive slices and corresponding PLs in a self-supervised manner. In the test stage, such correspondences are utilized to propagate a single annotated slice to the other slices of a test volume. We demonstrate the effectiveness of our method on various medical image segmentation tasks with different datasets, showing better generalizability across different organs, modalities, and modals. Code is available at \url{https://github.com/adlsn/Sli2Volplus}
Spectral U-Net: Enhancing Medical Image Segmentation via Spectral Decomposition
Sep 13, 2024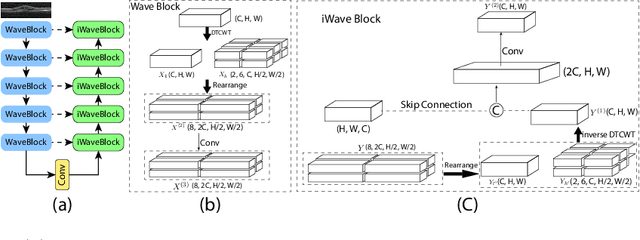


Abstract:This paper introduces Spectral U-Net, a novel deep learning network based on spectral decomposition, by exploiting Dual Tree Complex Wavelet Transform (DTCWT) for down-sampling and inverse Dual Tree Complex Wavelet Transform (iDTCWT) for up-sampling. We devise the corresponding Wave-Block and iWave-Block, integrated into the U-Net architecture, aiming at mitigating information loss during down-sampling and enhancing detail reconstruction during up-sampling. In the encoder, we first decompose the feature map into high and low-frequency components using DTCWT, enabling down-sampling while mitigating information loss. In the decoder, we utilize iDTCWT to reconstruct higher-resolution feature maps from down-sampled features. Evaluations on the Retina Fluid, Brain Tumor, and Liver Tumor segmentation datasets with the nnU-Net framework demonstrate the superiority of the proposed Spectral U-Net.
Denoising Diffusions in Latent Space for Medical Image Segmentation
Jul 17, 2024Abstract:Diffusion models (DPMs) have demonstrated remarkable performance in image generation, often times outperforming other generative models. Since their introduction, the powerful noise-to-image denoising pipeline has been extended to various discriminative tasks, including image segmentation. In case of medical imaging, often times the images are large 3D scans, where segmenting one image using DPMs become extremely inefficient due to large memory consumption and time consuming iterative sampling process. In this work, we propose a novel conditional generative modeling framework (LDSeg) that performs diffusion in latent space for medical image segmentation. Our proposed framework leverages the learned inherent low-dimensional latent distribution of the target object shapes and source image embeddings. The conditional diffusion in latent space not only ensures accurate n-D image segmentation for multi-label objects, but also mitigates the major underlying problems of the traditional DPM based segmentation: (1) large memory consumption, (2) time consuming sampling process and (3) unnatural noise injection in forward/reverse process. LDSeg achieved state-of-the-art segmentation accuracy on three medical image datasets with different imaging modalities. Furthermore, we show that our proposed model is significantly more robust to noises, compared to the traditional deterministic segmentation models, which can be potential in solving the domain shift problems in the medical imaging domain. Codes are available at: https://github.com/LDSeg/LDSeg.
Surf-CDM: Score-Based Surface Cold-Diffusion Model For Medical Image Segmentation
Dec 19, 2023Abstract:Diffusion models have shown impressive performance for image generation, often times outperforming other generative models. Since their introduction, researchers have extended the powerful noise-to-image denoising pipeline to discriminative tasks, including image segmentation. In this work we propose a conditional score-based generative modeling framework for medical image segmentation which relies on a parametric surface representation for the segmentation masks. The surface re-parameterization allows the direct application of standard diffusion theory, as opposed to when the mask is represented as a binary mask. Moreover, we adapted an extended variant of the diffusion technique known as the "cold-diffusion" where the diffusion model can be constructed with deterministic perturbations instead of Gaussian noise, which facilitates significantly faster convergence in the reverse diffusion. We evaluated our method on the segmentation of the left ventricle from 65 transthoracic echocardiogram videos (2230 echo image frames) and compared its performance to the most popular and widely used image segmentation models. Our proposed model not only outperformed the compared methods in terms of segmentation accuracy, but also showed potential in estimating segmentation uncertainties for further downstream analyses due to its inherent generative nature.
U-Net v2: Rethinking the Skip Connections of U-Net for Medical Image Segmentation
Nov 29, 2023



Abstract:In this paper, we introduce U-Net v2, a new robust and efficient U-Net variant for medical image segmentation. It aims to augment the infusion of semantic information into low-level features while simultaneously refining high-level features with finer details. For an input image, we begin by extracting multi-level features with a deep neural network encoder. Next, we enhance the feature map of each level by infusing semantic information from higher-level features and integrating finer details from lower-level features through Hadamard product. Our novel skip connections empower features of all the levels with enriched semantic characteristics and intricate details. The improved features are subsequently transmitted to the decoder for further processing and segmentation. Our method can be seamlessly integrated into any Encoder-Decoder network. We evaluate our method on several public medical image segmentation datasets for skin lesion segmentation and polyp segmentation, and the experimental results demonstrate the segmentation accuracy of our new method over state-of-the-art methods, while preserving memory and computational efficiency. Code is available at: https://github.com/yaoppeng/U-Net\_v2
PHG-Net: Persistent Homology Guided Medical Image Classification
Nov 28, 2023



Abstract:Modern deep neural networks have achieved great successes in medical image analysis. However, the features captured by convolutional neural networks (CNNs) or Transformers tend to be optimized for pixel intensities and neglect key anatomical structures such as connected components and loops. In this paper, we propose a persistent homology guided approach (PHG-Net) that explores topological features of objects for medical image classification. For an input image, we first compute its cubical persistence diagram and extract topological features into a vector representation using a small neural network (called the PH module). The extracted topological features are then incorporated into the feature map generated by CNN or Transformer for feature fusion. The PH module is lightweight and capable of integrating topological features into any CNN or Transformer architectures in an end-to-end fashion. We evaluate our PHG-Net on three public datasets and demonstrate its considerable improvements on the target classification tasks over state-of-the-art methods.
Trust, but Verify: Robust Image Segmentation using Deep Learning
Oct 29, 2023
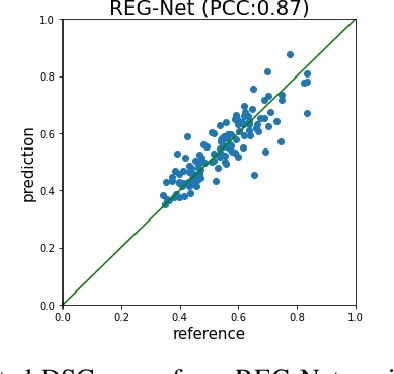
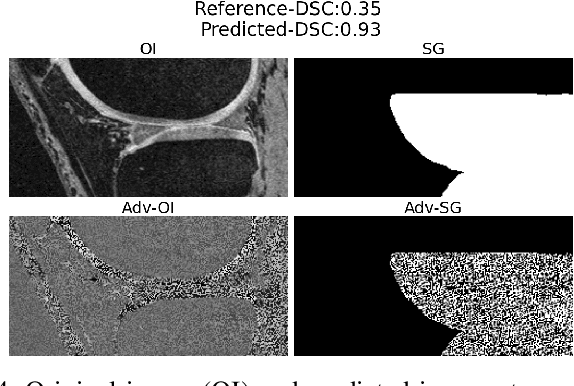
Abstract:We describe a method for verifying the output of a deep neural network for medical image segmentation that is robust to several classes of random as well as worst-case perturbations i.e. adversarial attacks. This method is based on a general approach recently developed by the authors called "Trust, but Verify" wherein an auxiliary verification network produces predictions about certain masked features in the input image using the segmentation as an input. A well-designed auxiliary network will produce high-quality predictions when the input segmentations are accurate, but will produce low-quality predictions when the segmentations are incorrect. Checking the predictions of such a network with the original image allows us to detect bad segmentations. However, to ensure the verification method is truly robust, we need a method for checking the quality of the predictions that does not itself rely on a black-box neural network. Indeed, we show that previous methods for segmentation evaluation that do use deep neural regression networks are vulnerable to false negatives i.e. can inaccurately label bad segmentations as good. We describe the design of a verification network that avoids such vulnerability and present results to demonstrate its robustness compared to previous methods.
Provable Multi-instance Deep AUC Maximization with Stochastic Pooling
May 18, 2023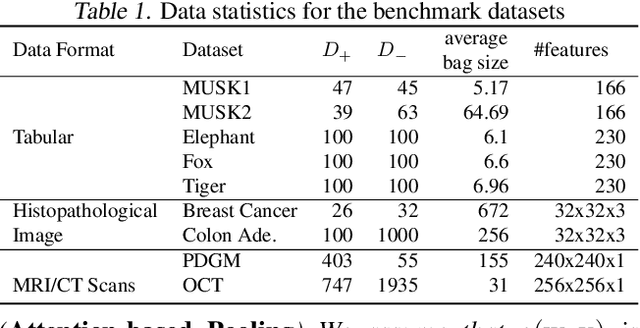
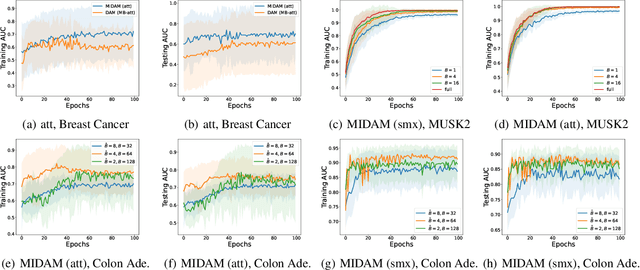

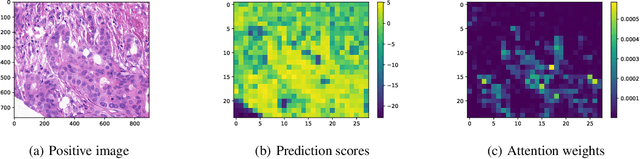
Abstract:This paper considers a novel application of deep AUC maximization (DAM) for multi-instance learning (MIL), in which a single class label is assigned to a bag of instances (e.g., multiple 2D slices of a CT scan for a patient). We address a neglected yet non-negligible computational challenge of MIL in the context of DAM, i.e., bag size is too large to be loaded into {GPU} memory for backpropagation, which is required by the standard pooling methods of MIL. To tackle this challenge, we propose variance-reduced stochastic pooling methods in the spirit of stochastic optimization by formulating the loss function over the pooled prediction as a multi-level compositional function. By synthesizing techniques from stochastic compositional optimization and non-convex min-max optimization, we propose a unified and provable muli-instance DAM (MIDAM) algorithm with stochastic smoothed-max pooling or stochastic attention-based pooling, which only samples a few instances for each bag to compute a stochastic gradient estimator and to update the model parameter. We establish a similar convergence rate of the proposed MIDAM algorithm as the state-of-the-art DAM algorithms. Our extensive experiments on conventional MIL datasets and medical datasets demonstrate the superiority of our MIDAM algorithm.
Automated segmentation of choroidal layers from 3-dimensional macular optical coherence tomography scans
Mar 11, 2021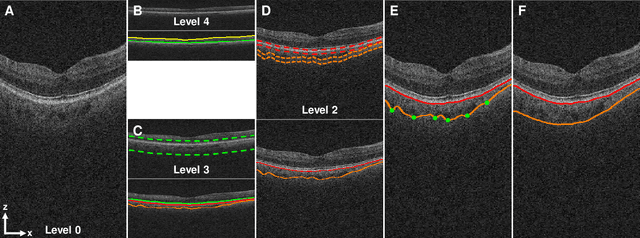
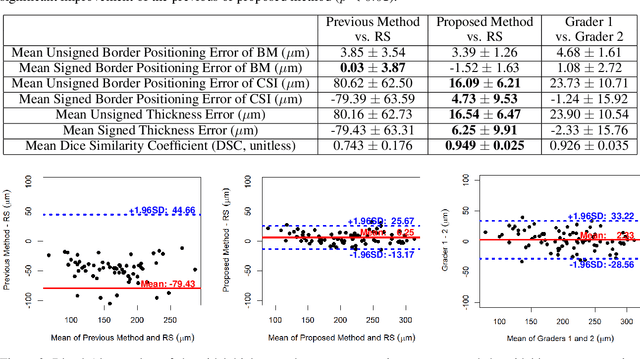
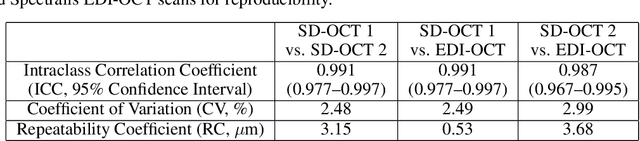
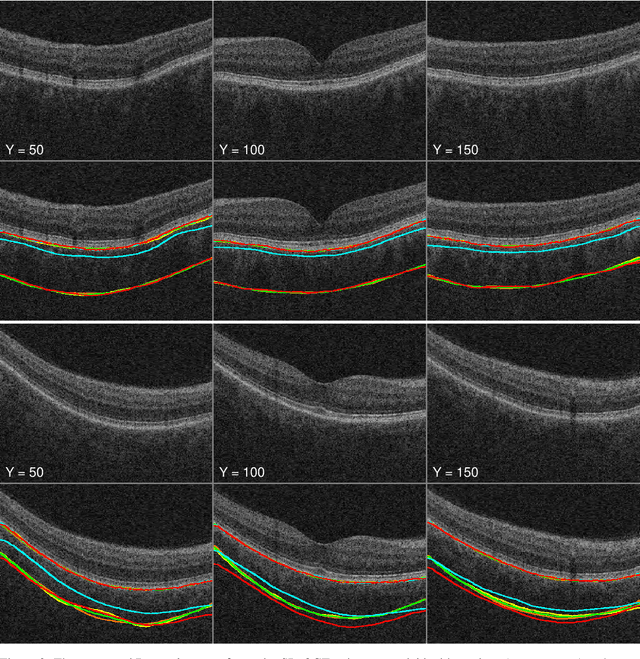
Abstract:Background: Changes in choroidal thickness are associated with various ocular diseases and the choroid can be imaged using spectral-domain optical coherence tomography (SDOCT) and enhanced depth imaging OCT (EDIOCT). New Method: Eighty macular SDOCT volumes from 80 patients were obtained using the Zeiss Cirrus machine. Eleven additional control subjects had two Cirrus scans done in one visit along with EDIOCT using the Heidelberg Spectralis machine. To automatically segment choroidal layers from the OCT volumes, our graph-theoretic approach was utilized. The segmentation results were compared with reference standards from two graders, and the accuracy of automated segmentation was calculated using unsigned to signed border positioning thickness errors and Dice similarity coefficient (DSC). The repeatability and reproducibility of our choroidal thicknesses were determined by intraclass correlation coefficient (ICC), coefficient of variation (CV), and repeatability coefficient (RC). Results: The mean unsigned to signed border positioning errors for the choroidal inner and outer surfaces are 3.39plusminus1.26microns (mean plusminus SD) to minus1.52 plusminus 1.63microns and 16.09 plusminus 6.21microns to 4.73 plusminus 9.53microns, respectively. The mean unsigned to signed choroidal thickness errors are 16.54 plusminus 6.47microns to 6.25 plusminus 9.91microns, and the mean DSC is 0.949 plusminus 0.025. The ICC (95% CI), CV, RC values are 0.991 (0.977 to 0.997), 2.48%, 3.15microns for the repeatability and 0.991 (0.977 to 0.997), 2.49%, 0.53microns for the reproducibility studies, respectively. Comparison with Existing Method(s): The proposed method outperformed our previous method using choroidal vessel segmentation and inter-grader variability. Conclusions: This automated segmentation method can reliably measure choroidal thickness using different OCT platforms.
Robust Deep AUC Maximization: A New Surrogate Loss and Empirical Studies on Medical Image Classification
Dec 06, 2020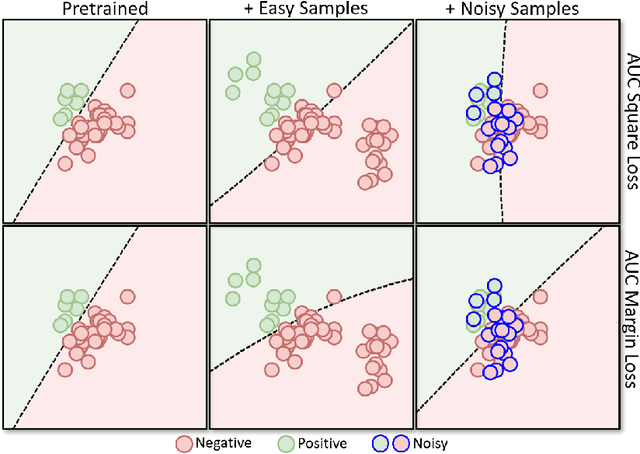
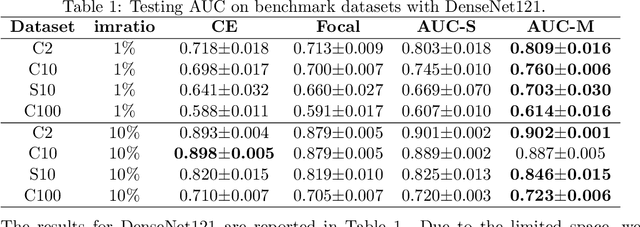
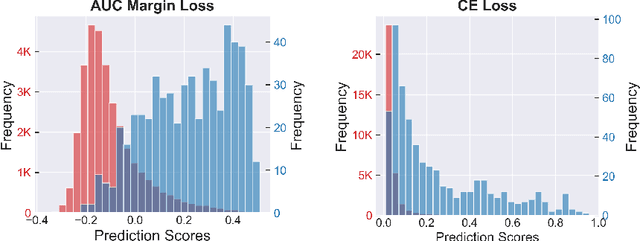
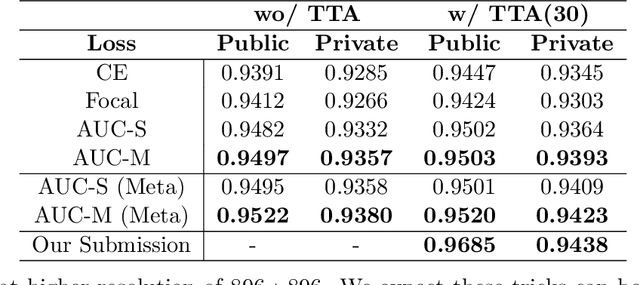
Abstract:Deep AUC Maximization (DAM) is a paradigm for learning a deep neural network by maximizing the AUC score of the model on a dataset. Most previous works of AUC maximization focus on the perspective of optimization by designing efficient stochastic algorithms, and studies on generalization performance of DAM on difficult tasks are missing. In this work, we aim to make DAM more practical for interesting real-world applications (e.g., medical image classification). First, we propose a new margin-based surrogate loss function for the AUC score (named as the AUC margin loss). It is more robust than the commonly used AUC square loss, while enjoying the same advantage in terms of large-scale stochastic optimization. Second, we conduct empirical studies of our DAM method on difficult medical image classification tasks, namely classification of chest x-ray images for identifying many threatening diseases and classification of images of skin lesions for identifying melanoma. Our DAM method has achieved great success on these difficult tasks, i.e., the 1st place on Stanford CheXpert competition (by the paper submission date) and Top 1% rank (rank 33 out of 3314 teams) on Kaggle 2020 Melanoma classification competition. We also conduct extensive ablation studies to demonstrate the advantages of the new AUC margin loss over the AUC square loss on benchmark datasets. To the best of our knowledge, this is the first work that makes DAM succeed on large-scale medical image datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge