Xiaodong Wu
SafeLLM: Unlearning Harmful Outputs from Large Language Models against Jailbreak Attacks
Aug 21, 2025Abstract:Jailbreak attacks pose a serious threat to the safety of Large Language Models (LLMs) by crafting adversarial prompts that bypass alignment mechanisms, causing the models to produce harmful, restricted, or biased content. In this paper, we propose SafeLLM, a novel unlearning-based defense framework that unlearn the harmful knowledge from LLMs while preserving linguistic fluency and general capabilities. SafeLLM employs a three-stage pipeline: (1) dynamic unsafe output detection using a hybrid approach that integrates external classifiers with model-internal evaluations; (2) token-level harmful content tracing through feedforward network (FFN) activations to localize harmful knowledge; and (3) constrained optimization to suppress unsafe behavior without degrading overall model quality. SafeLLM achieves targeted and irreversible forgetting by identifying and neutralizing FFN substructures responsible for harmful generation pathways. Extensive experiments on prominent LLMs (Vicuna, LLaMA, and GPT-J) across multiple jailbreak benchmarks show that SafeLLM substantially reduces attack success rates while maintaining high general-purpose performance. Compared to standard defense methods such as supervised fine-tuning and direct preference optimization, SafeLLM offers stronger safety guarantees, more precise control over harmful behavior, and greater robustness to unseen attacks. Moreover, SafeLLM maintains the general performance after the harmful knowledge unlearned. These results highlight unlearning as a promising direction for scalable and effective LLM safety.
LIFBench: Evaluating the Instruction Following Performance and Stability of Large Language Models in Long-Context Scenarios
Nov 11, 2024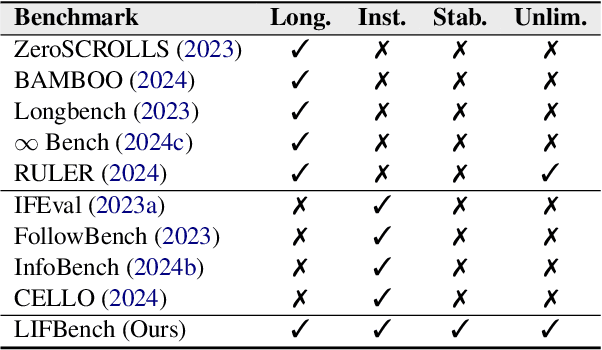
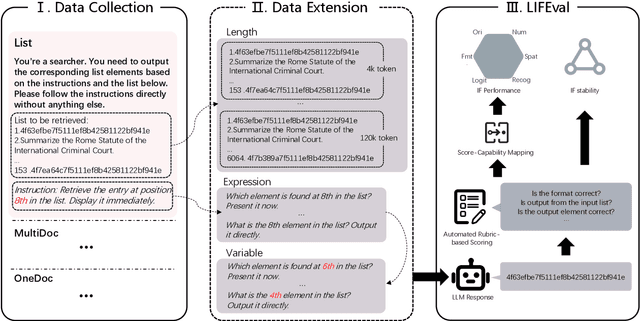
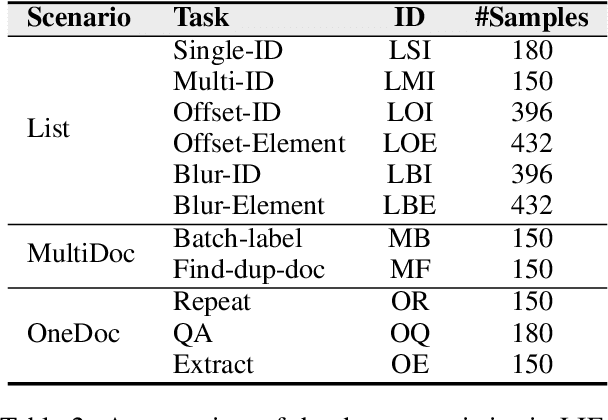
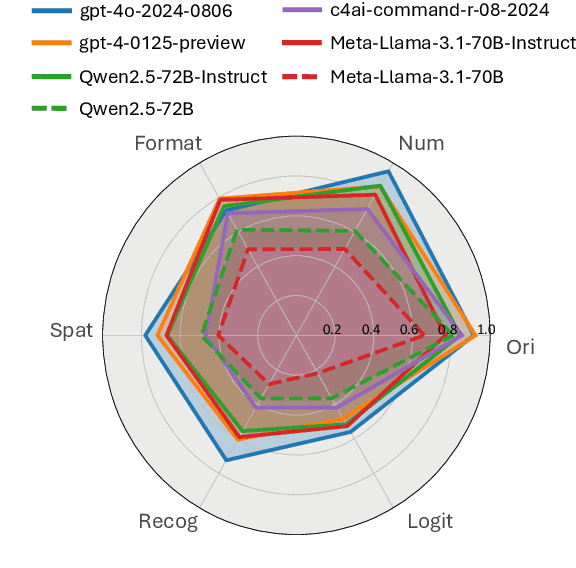
Abstract:As Large Language Models (LLMs) continue to advance in natural language processing (NLP), their ability to stably follow instructions in long-context inputs has become crucial for real-world applications. While existing benchmarks assess various LLM capabilities, they rarely focus on instruction-following in long-context scenarios or stability on different inputs. In response, we introduce the Long-context Instruction-Following Benchmark (LIFBench), a scalable dataset designed to evaluate LLMs' instruction-following capabilities and stability across long contexts. LIFBench comprises three long-context scenarios and eleven diverse tasks, supported by 2,766 instructions generated through an automated expansion method across three dimensions: length, expression, and variables. For evaluation, we propose LIFEval, a rubric-based assessment framework that provides precise, automated scoring of complex LLM responses without relying on LLM-assisted evaluations or human judgments. This approach facilitates a comprehensive analysis of model performance and stability across various perspectives. We conduct extensive experiments on 20 notable LLMs across six length intervals, analyzing their instruction-following capabilities and stability. Our work contributes LIFBench and LIFEval as robust tools for assessing LLM performance in complex, long-context settings, providing insights that can inform future LLM development.
Denoising Diffusions in Latent Space for Medical Image Segmentation
Jul 17, 2024Abstract:Diffusion models (DPMs) have demonstrated remarkable performance in image generation, often times outperforming other generative models. Since their introduction, the powerful noise-to-image denoising pipeline has been extended to various discriminative tasks, including image segmentation. In case of medical imaging, often times the images are large 3D scans, where segmenting one image using DPMs become extremely inefficient due to large memory consumption and time consuming iterative sampling process. In this work, we propose a novel conditional generative modeling framework (LDSeg) that performs diffusion in latent space for medical image segmentation. Our proposed framework leverages the learned inherent low-dimensional latent distribution of the target object shapes and source image embeddings. The conditional diffusion in latent space not only ensures accurate n-D image segmentation for multi-label objects, but also mitigates the major underlying problems of the traditional DPM based segmentation: (1) large memory consumption, (2) time consuming sampling process and (3) unnatural noise injection in forward/reverse process. LDSeg achieved state-of-the-art segmentation accuracy on three medical image datasets with different imaging modalities. Furthermore, we show that our proposed model is significantly more robust to noises, compared to the traditional deterministic segmentation models, which can be potential in solving the domain shift problems in the medical imaging domain. Codes are available at: https://github.com/LDSeg/LDSeg.
Towards unlocking the mystery of adversarial fragility of neural networks
Jun 23, 2024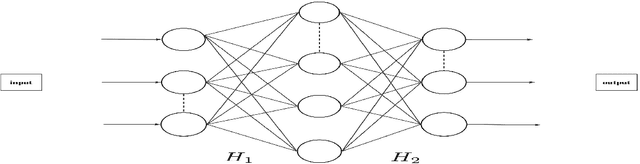
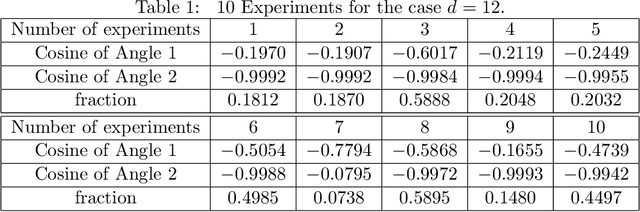
Abstract:In this paper, we study the adversarial robustness of deep neural networks for classification tasks. We look at the smallest magnitude of possible additive perturbations that can change the output of a classification algorithm. We provide a matrix-theoretic explanation of the adversarial fragility of deep neural network for classification. In particular, our theoretical results show that neural network's adversarial robustness can degrade as the input dimension $d$ increases. Analytically we show that neural networks' adversarial robustness can be only $1/\sqrt{d}$ of the best possible adversarial robustness. Our matrix-theoretic explanation is consistent with an earlier information-theoretic feature-compression-based explanation for the adversarial fragility of neural networks.
An Improved Empirical Fisher Approximation for Natural Gradient Descent
Jun 10, 2024Abstract:Approximate Natural Gradient Descent (NGD) methods are an important family of optimisers for deep learning models, which use approximate Fisher information matrices to pre-condition gradients during training. The empirical Fisher (EF) method approximates the Fisher information matrix empirically by reusing the per-sample gradients collected during back-propagation. Despite its ease of implementation, the EF approximation has its theoretical and practical limitations. This paper first investigates the inversely-scaled projection issue of EF, which is shown to be a major cause of the poor empirical approximation quality. An improved empirical Fisher (iEF) method, motivated as a generalised NGD method from a loss reduction perspective, is proposed to address this issue, meanwhile retaining the practical convenience of EF. The exact iEF and EF methods are experimentally evaluated using practical deep learning setups, including widely-used setups for parameter-efficient fine-tuning of pre-trained models (T5-base with LoRA and Prompt-Tuning on GLUE tasks, and ViT with LoRA for CIFAR100). Optimisation experiments show that applying exact iEF as an optimiser provides strong convergence and generalisation. It achieves the best test performance and the lowest training loss for majority of the tasks, even when compared with well-tuned AdamW/Adafactor baselines. Additionally, under a novel empirical evaluation framework, the proposed iEF method shows consistently better approximation quality to the exact Natural Gradient updates than both EF and the more expensive sampled Fisher (SF). Further investigation also shows that the superior approximation quality of iEF is robust to damping across tasks and training stages. Improving existing approximate NGD optimisers with iEF is expected to lead to better convergence ability and stronger robustness to choice of damping.
Manipulating Predictions over Discrete Inputs in Machine Teaching
Jan 31, 2024Abstract:Machine teaching often involves the creation of an optimal (typically minimal) dataset to help a model (referred to as the `student') achieve specific goals given by a teacher. While abundant in the continuous domain, the studies on the effectiveness of machine teaching in the discrete domain are relatively limited. This paper focuses on machine teaching in the discrete domain, specifically on manipulating student models' predictions based on the goals of teachers via changing the training data efficiently. We formulate this task as a combinatorial optimization problem and solve it by proposing an iterative searching algorithm. Our algorithm demonstrates significant numerical merit in the scenarios where a teacher attempts at correcting erroneous predictions to improve the student's models, or maliciously manipulating the model to misclassify some specific samples to the target class aligned with his personal profits. Experimental results show that our proposed algorithm can have superior performance in effectively and efficiently manipulating the predictions of the model, surpassing conventional baselines.
Diagnosis Of Takotsubo Syndrome By Robust Feature Selection From The Complex Latent Space Of DL-based Segmentation Network
Dec 19, 2023Abstract:Researchers have shown significant correlations among segmented objects in various medical imaging modalities and disease related pathologies. Several studies showed that using hand crafted features for disease prediction neglects the immense possibility to use latent features from deep learning (DL) models which may reduce the overall accuracy of differential diagnosis. However, directly using classification or segmentation models on medical to learn latent features opt out robust feature selection and may lead to overfitting. To fill this gap, we propose a novel feature selection technique using the latent space of a segmentation model that can aid diagnosis. We evaluated our method in differentiating a rare cardiac disease: Takotsubo Syndrome (TTS) from the ST elevation myocardial infarction (STEMI) using echocardiogram videos (echo). TTS can mimic clinical features of STEMI in echo and extremely hard to distinguish. Our approach shows promising results in differential diagnosis of TTS with 82% diagnosis accuracy beating the previous state-of-the-art (SOTA) approach. Moreover, the robust feature selection technique using LASSO algorithm shows great potential in reducing the redundant features and creates a robust pipeline for short- and long-term disease prognoses in the downstream analysis.
Surf-CDM: Score-Based Surface Cold-Diffusion Model For Medical Image Segmentation
Dec 19, 2023Abstract:Diffusion models have shown impressive performance for image generation, often times outperforming other generative models. Since their introduction, researchers have extended the powerful noise-to-image denoising pipeline to discriminative tasks, including image segmentation. In this work we propose a conditional score-based generative modeling framework for medical image segmentation which relies on a parametric surface representation for the segmentation masks. The surface re-parameterization allows the direct application of standard diffusion theory, as opposed to when the mask is represented as a binary mask. Moreover, we adapted an extended variant of the diffusion technique known as the "cold-diffusion" where the diffusion model can be constructed with deterministic perturbations instead of Gaussian noise, which facilitates significantly faster convergence in the reverse diffusion. We evaluated our method on the segmentation of the left ventricle from 65 transthoracic echocardiogram videos (2230 echo image frames) and compared its performance to the most popular and widely used image segmentation models. Our proposed model not only outperformed the compared methods in terms of segmentation accuracy, but also showed potential in estimating segmentation uncertainties for further downstream analyses due to its inherent generative nature.
gcDLSeg: Integrating Graph-cut into Deep Learning for Binary Semantic Segmentation
Dec 07, 2023



Abstract:Binary semantic segmentation in computer vision is a fundamental problem. As a model-based segmentation method, the graph-cut approach was one of the most successful binary segmentation methods thanks to its global optimality guarantee of the solutions and its practical polynomial-time complexity. Recently, many deep learning (DL) based methods have been developed for this task and yielded remarkable performance, resulting in a paradigm shift in this field. To combine the strengths of both approaches, we propose in this study to integrate the graph-cut approach into a deep learning network for end-to-end learning. Unfortunately, backward propagation through the graph-cut module in the DL network is challenging due to the combinatorial nature of the graph-cut algorithm. To tackle this challenge, we propose a novel residual graph-cut loss and a quasi-residual connection, enabling the backward propagation of the gradients of the residual graph-cut loss for effective feature learning guided by the graph-cut segmentation model. In the inference phase, globally optimal segmentation is achieved with respect to the graph-cut energy defined on the optimized image features learned from DL networks. Experiments on the public AZH chronic wound data set and the pancreas cancer data set from the medical segmentation decathlon (MSD) demonstrated promising segmentation accuracy, and improved robustness against adversarial attacks.
Trust, but Verify: Robust Image Segmentation using Deep Learning
Oct 29, 2023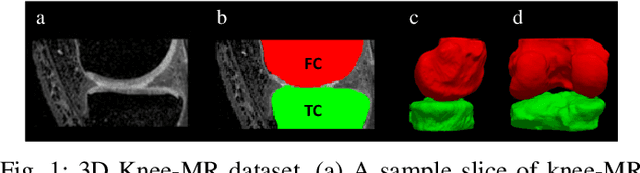
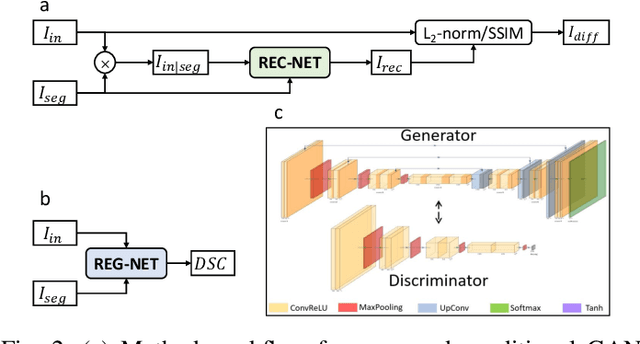
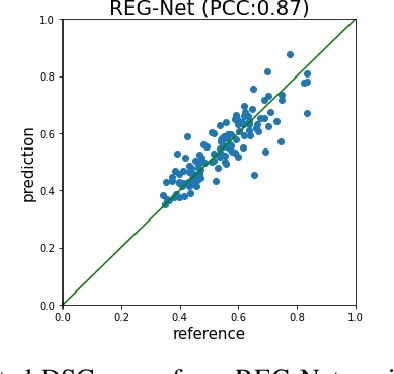
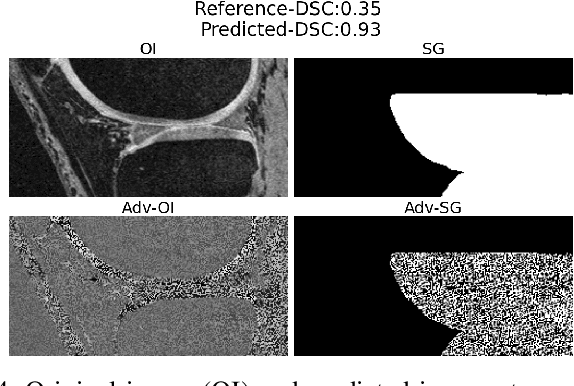
Abstract:We describe a method for verifying the output of a deep neural network for medical image segmentation that is robust to several classes of random as well as worst-case perturbations i.e. adversarial attacks. This method is based on a general approach recently developed by the authors called "Trust, but Verify" wherein an auxiliary verification network produces predictions about certain masked features in the input image using the segmentation as an input. A well-designed auxiliary network will produce high-quality predictions when the input segmentations are accurate, but will produce low-quality predictions when the segmentations are incorrect. Checking the predictions of such a network with the original image allows us to detect bad segmentations. However, to ensure the verification method is truly robust, we need a method for checking the quality of the predictions that does not itself rely on a black-box neural network. Indeed, we show that previous methods for segmentation evaluation that do use deep neural regression networks are vulnerable to false negatives i.e. can inaccurately label bad segmentations as good. We describe the design of a verification network that avoids such vulnerability and present results to demonstrate its robustness compared to previous methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge