Minhao Wang
Beyond Semantic Understanding: Preserving Collaborative Frequency Components in LLM-based Recommendation
Aug 14, 2025Abstract:Recommender systems in concert with Large Language Models (LLMs) present promising avenues for generating semantically-informed recommendations. However, LLM-based recommenders exhibit a tendency to overemphasize semantic correlations within users' interaction history. When taking pretrained collaborative ID embeddings as input, LLM-based recommenders progressively weaken the inherent collaborative signals as the embeddings propagate through LLM backbones layer by layer, as opposed to traditional Transformer-based sequential models in which collaborative signals are typically preserved or even enhanced for state-of-the-art performance. To address this limitation, we introduce FreLLM4Rec, an approach designed to balance semantic and collaborative information from a spectral perspective. Item embeddings that incorporate both semantic and collaborative information are first purified using a Global Graph Low-Pass Filter (G-LPF) to preliminarily remove irrelevant high-frequency noise. Temporal Frequency Modulation (TFM) then actively preserves collaborative signal layer by layer. Note that the collaborative preservation capability of TFM is theoretically guaranteed by establishing a connection between the optimal but hard-to-implement local graph fourier filters and the suboptimal yet computationally efficient frequency-domain filters. Extensive experiments on four benchmark datasets demonstrate that FreLLM4Rec successfully mitigates collaborative signal attenuation and achieves competitive performance, with improvements of up to 8.00\% in NDCG@10 over the best baseline. Our findings provide insights into how LLMs process collaborative information and offer a principled approach for improving LLM-based recommendation systems.
LIFBench: Evaluating the Instruction Following Performance and Stability of Large Language Models in Long-Context Scenarios
Nov 11, 2024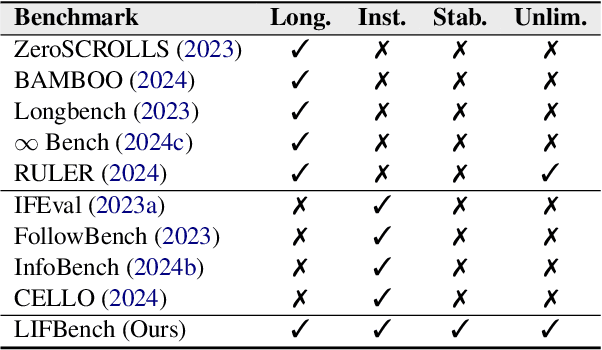
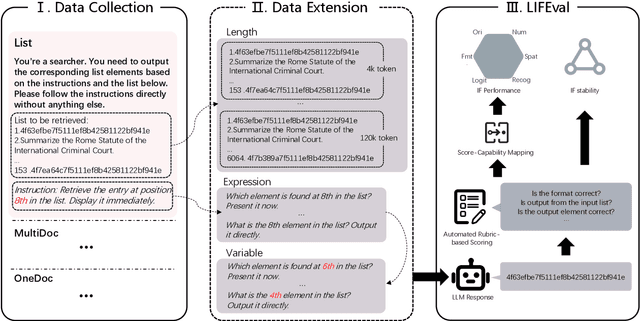
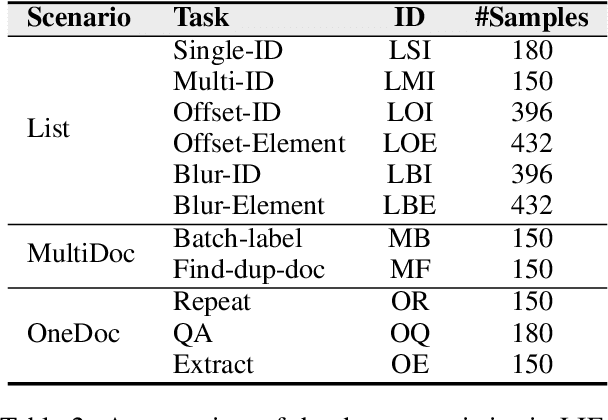
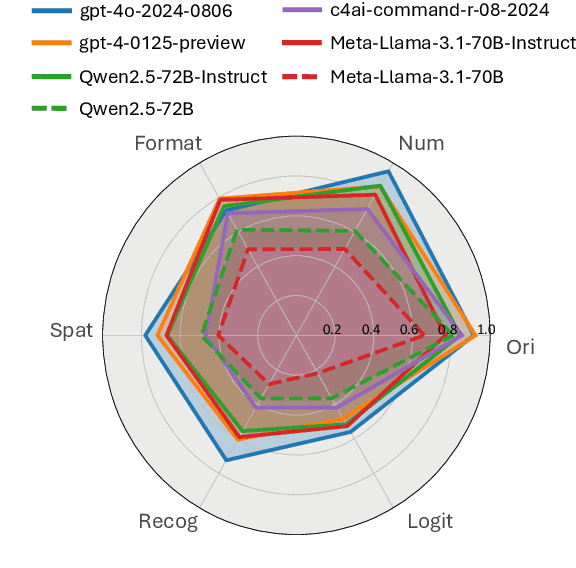
Abstract:As Large Language Models (LLMs) continue to advance in natural language processing (NLP), their ability to stably follow instructions in long-context inputs has become crucial for real-world applications. While existing benchmarks assess various LLM capabilities, they rarely focus on instruction-following in long-context scenarios or stability on different inputs. In response, we introduce the Long-context Instruction-Following Benchmark (LIFBench), a scalable dataset designed to evaluate LLMs' instruction-following capabilities and stability across long contexts. LIFBench comprises three long-context scenarios and eleven diverse tasks, supported by 2,766 instructions generated through an automated expansion method across three dimensions: length, expression, and variables. For evaluation, we propose LIFEval, a rubric-based assessment framework that provides precise, automated scoring of complex LLM responses without relying on LLM-assisted evaluations or human judgments. This approach facilitates a comprehensive analysis of model performance and stability across various perspectives. We conduct extensive experiments on 20 notable LLMs across six length intervals, analyzing their instruction-following capabilities and stability. Our work contributes LIFBench and LIFEval as robust tools for assessing LLM performance in complex, long-context settings, providing insights that can inform future LLM development.
Robust Weakly Supervised Learning for COVID-19 Recognition Using Multi-Center CT Images
Dec 09, 2021



Abstract:The world is currently experiencing an ongoing pandemic of an infectious disease named coronavirus disease 2019 (i.e., COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Computed Tomography (CT) plays an important role in assessing the severity of the infection and can also be used to identify those symptomatic and asymptomatic COVID-19 carriers. With a surge of the cumulative number of COVID-19 patients, radiologists are increasingly stressed to examine the CT scans manually. Therefore, an automated 3D CT scan recognition tool is highly in demand since the manual analysis is time-consuming for radiologists and their fatigue can cause possible misjudgment. However, due to various technical specifications of CT scanners located in different hospitals, the appearance of CT images can be significantly different leading to the failure of many automated image recognition approaches. The multi-domain shift problem for the multi-center and multi-scanner studies is therefore nontrivial that is also crucial for a dependable recognition and critical for reproducible and objective diagnosis and prognosis. In this paper, we proposed a COVID-19 CT scan recognition model namely coronavirus information fusion and diagnosis network (CIFD-Net) that can efficiently handle the multi-domain shift problem via a new robust weakly supervised learning paradigm. Our model can resolve the problem of different appearance in CT scan images reliably and efficiently while attaining higher accuracy compared to other state-of-the-art methods.
Weakly Supervised Deep Learning for COVID-19 Infection Detection and Classification from CT Images
Apr 14, 2020



Abstract:An outbreak of a novel coronavirus disease (i.e., COVID-19) has been recorded in Wuhan, China since late December 2019, which subsequently became pandemic around the world. Although COVID-19 is an acutely treated disease, it can also be fatal with a risk of fatality of 4.03% in China and the highest of 13.04% in Algeria and 12.67% Italy (as of 8th April 2020). The onset of serious illness may result in death as a consequence of substantial alveolar damage and progressive respiratory failure. Although laboratory testing, e.g., using reverse transcription polymerase chain reaction (RT-PCR), is the golden standard for clinical diagnosis, the tests may produce false negatives. Moreover, under the pandemic situation, shortage of RT-PCR testing resources may also delay the following clinical decision and treatment. Under such circumstances, chest CT imaging has become a valuable tool for both diagnosis and prognosis of COVID-19 patients. In this study, we propose a weakly supervised deep learning strategy for detecting and classifying COVID-19 infection from CT images. The proposed method can minimise the requirements of manual labelling of CT images but still be able to obtain accurate infection detection and distinguish COVID-19 from non-COVID-19 cases. Based on the promising results obtained qualitatively and quantitatively, we can envisage a wide deployment of our developed technique in large-scale clinical studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge