Chengjia Wang
REF-VC: Robust, Expressive and Fast Zero-Shot Voice Conversion with Diffusion Transformers
Aug 07, 2025Abstract:In real-world voice conversion applications, environmental noise in source speech and user demands for expressive output pose critical challenges. Traditional ASR-based methods ensure noise robustness but suppress prosody, while SSL-based models improve expressiveness but suffer from timbre leakage and noise sensitivity. This paper proposes REF-VC, a noise-robust expressive voice conversion system. Key innovations include: (1) A random erasing strategy to mitigate the information redundancy inherent in SSL feature, enhancing noise robustness and expressiveness; (2) Implicit alignment inspired by E2TTS to suppress non-essential feature reconstruction; (3) Integration of Shortcut Models to accelerate flow matching inference, significantly reducing to 4 steps. Experimental results demonstrate that our model outperforms baselines such as Seed-VC in zero-shot scenarios on the noisy set, while also performing comparably to Seed-VC on the clean set. In addition, REF-VC can be compatible with singing voice conversion within one model.
A Novel Coronary Artery Registration Method Based on Super-pixel Particle Swarm Optimization
May 30, 2025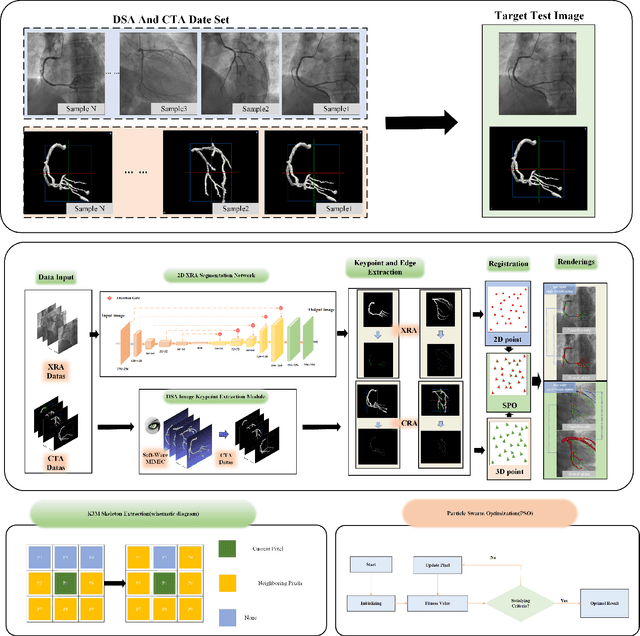
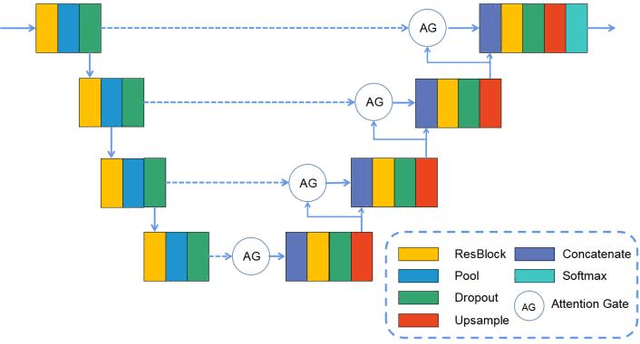
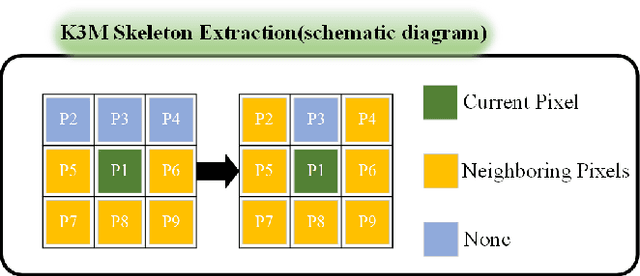
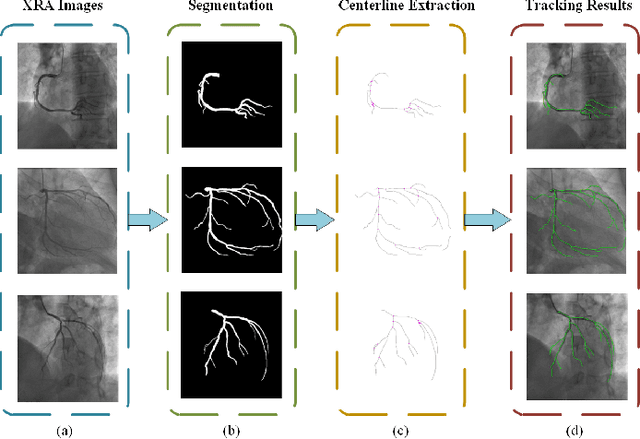
Abstract:Percutaneous Coronary Intervention (PCI) is a minimally invasive procedure that improves coronary blood flow and treats coronary artery disease. Although PCI typically requires 2D X-ray angiography (XRA) to guide catheter placement at real-time, computed tomography angiography (CTA) may substantially improve PCI by providing precise information of 3D vascular anatomy and status. To leverage real-time XRA and detailed 3D CTA anatomy for PCI, accurate multimodal image registration of XRA and CTA is required, to guide the procedure and avoid complications. This is a challenging process as it requires registration of images from different geometrical modalities (2D -> 3D and vice versa), with variations in contrast and noise levels. In this paper, we propose a novel multimodal coronary artery image registration method based on a swarm optimization algorithm, which effectively addresses challenges such as large deformations, low contrast, and noise across these imaging modalities. Our algorithm consists of two main modules: 1) preprocessing of XRA and CTA images separately, and 2) a registration module based on feature extraction using the Steger and Superpixel Particle Swarm Optimization algorithms. Our technique was evaluated on a pilot dataset of 28 pairs of XRA and CTA images from 10 patients who underwent PCI. The algorithm was compared with four state-of-the-art (SOTA) methods in terms of registration accuracy, robustness, and efficiency. Our method outperformed the selected SOTA baselines in all aspects. Experimental results demonstrate the significant effectiveness of our algorithm, surpassing the previous benchmarks and proposes a novel clinical approach that can potentially have merit for improving patient outcomes in coronary artery disease.
Scale Invariance of Graph Neural Networks
Nov 28, 2024



Abstract:We address two fundamental challenges in Graph Neural Networks (GNNs): (1) the lack of theoretical support for invariance learning, a critical property in image processing, and (2) the absence of a unified model capable of excelling on both homophilic and heterophilic graph datasets. To tackle these issues, we establish and prove scale invariance in graphs, extending this key property to graph learning, and validate it through experiments on real-world datasets. Leveraging directed multi-scaled graphs and an adaptive self-loop strategy, we propose ScaleNet, a unified network architecture that achieves state-of-the-art performance across four homophilic and two heterophilic benchmark datasets. Furthermore, we show that through graph transformation based on scale invariance, uniform weights can replace computationally expensive edge weights in digraph inception networks while maintaining or improving performance. For another popular GNN approach to digraphs, we demonstrate the equivalence between Hermitian Laplacian methods and GraphSAGE with incidence normalization. ScaleNet bridges the gap between homophilic and heterophilic graph learning, offering both theoretical insights into scale invariance and practical advancements in unified graph learning. Our implementation is publicly available at https://github.com/Qin87/ScaleNet/tree/Aug23.
ScaleNet: Scale Invariance Learning in Directed Graphs
Nov 13, 2024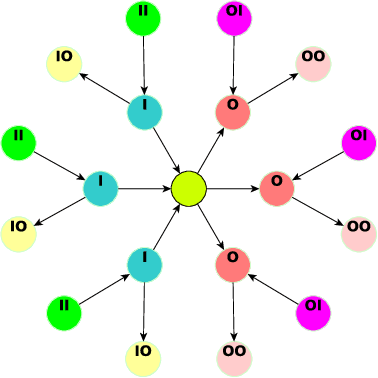
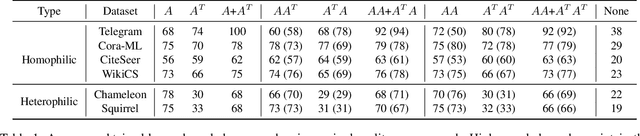
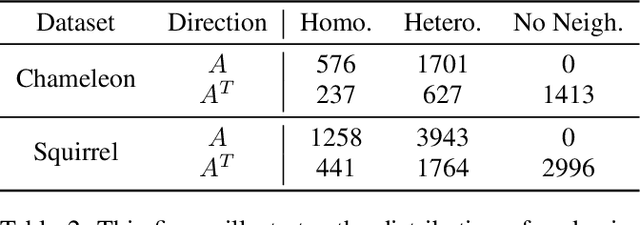
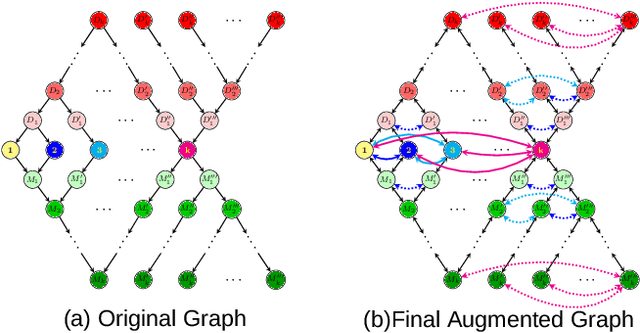
Abstract:Graph Neural Networks (GNNs) have advanced relational data analysis but lack invariance learning techniques common in image classification. In node classification with GNNs, it is actually the ego-graph of the center node that is classified. This research extends the scale invariance concept to node classification by drawing an analogy to image processing: just as scale invariance being used in image classification to capture multi-scale features, we propose the concept of ``scaled ego-graphs''. Scaled ego-graphs generalize traditional ego-graphs by replacing undirected single-edges with ``scaled-edges'', which are ordered sequences of multiple directed edges. We empirically assess the performance of the proposed scale invariance in graphs on seven benchmark datasets, across both homophilic and heterophilic structures. Our scale-invariance-based graph learning outperforms inception models derived from random walks by being simpler, faster, and more accurate. The scale invariance explains inception models' success on homophilic graphs and limitations on heterophilic graphs. To ensure applicability of inception model to heterophilic graphs as well, we further present ScaleNet, an architecture that leverages multi-scaled features. ScaleNet achieves state-of-the-art results on five out of seven datasets (four homophilic and one heterophilic) and matches top performance on the remaining two, demonstrating its excellent applicability. This represents a significant advance in graph learning, offering a unified framework that enhances node classification across various graph types. Our code is available at https://github.com/Qin87/ScaleNet/tree/July25.
SeLoRA: Self-Expanding Low-Rank Adaptation of Latent Diffusion Model for Medical Image Synthesis
Aug 13, 2024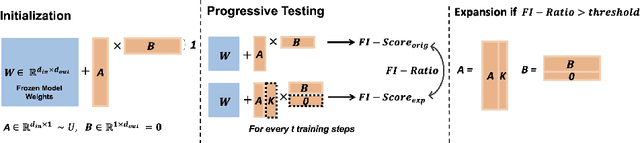
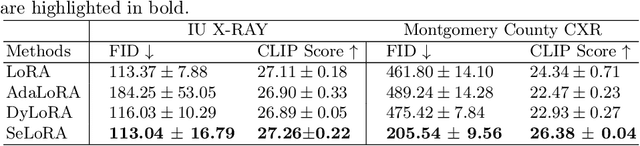
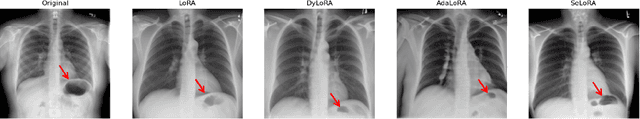

Abstract:The persistent challenge of medical image synthesis posed by the scarcity of annotated data and the need to synthesize `missing modalities' for multi-modal analysis, underscored the imperative development of effective synthesis methods. Recently, the combination of Low-Rank Adaptation (LoRA) with latent diffusion models (LDMs) has emerged as a viable approach for efficiently adapting pre-trained large language models, in the medical field. However, the direct application of LoRA assumes uniform ranking across all linear layers, overlooking the significance of different weight matrices, and leading to sub-optimal outcomes. Prior works on LoRA prioritize the reduction of trainable parameters, and there exists an opportunity to further tailor this adaptation process to the intricate demands of medical image synthesis. In response, we present SeLoRA, a Self-Expanding Low-Rank Adaptation Module, that dynamically expands its ranking across layers during training, strategically placing additional ranks on crucial layers, to allow the model to elevate synthesis quality where it matters most. The proposed method not only enables LDMs to fine-tune on medical data efficiently but also empowers the model to achieve improved image quality with minimal ranking. The code of our SeLoRA method is publicly available on https://anonymous.4open.science/r/SeLoRA-980D .
Explainable unsupervised multi-modal image registration using deep networks
Aug 03, 2023Abstract:Clinical decision making from magnetic resonance imaging (MRI) combines complementary information from multiple MRI sequences (defined as 'modalities'). MRI image registration aims to geometrically 'pair' diagnoses from different modalities, time points and slices. Both intra- and inter-modality MRI registration are essential components in clinical MRI settings. Further, an MRI image processing pipeline that can address both afine and non-rigid registration is critical, as both types of deformations may be occuring in real MRI data scenarios. Unlike image classification, explainability is not commonly addressed in image registration deep learning (DL) methods, as it is challenging to interpet model-data behaviours against transformation fields. To properly address this, we incorporate Grad-CAM-based explainability frameworks in each major component of our unsupervised multi-modal and multi-organ image registration DL methodology. We previously demonstrated that we were able to reach superior performance (against the current standard Syn method). In this work, we show that our DL model becomes fully explainable, setting the framework to generalise our approach on further medical imaging data.
Is attention all you need in medical image analysis? A review
Jul 24, 2023Abstract:Medical imaging is a key component in clinical diagnosis, treatment planning and clinical trial design, accounting for almost 90% of all healthcare data. CNNs achieved performance gains in medical image analysis (MIA) over the last years. CNNs can efficiently model local pixel interactions and be trained on small-scale MI data. The main disadvantage of typical CNN models is that they ignore global pixel relationships within images, which limits their generalisation ability to understand out-of-distribution data with different 'global' information. The recent progress of Artificial Intelligence gave rise to Transformers, which can learn global relationships from data. However, full Transformer models need to be trained on large-scale data and involve tremendous computational complexity. Attention and Transformer compartments (Transf/Attention) which can well maintain properties for modelling global relationships, have been proposed as lighter alternatives of full Transformers. Recently, there is an increasing trend to co-pollinate complementary local-global properties from CNN and Transf/Attention architectures, which led to a new era of hybrid models. The past years have witnessed substantial growth in hybrid CNN-Transf/Attention models across diverse MIA problems. In this systematic review, we survey existing hybrid CNN-Transf/Attention models, review and unravel key architectural designs, analyse breakthroughs, and evaluate current and future opportunities as well as challenges. We also introduced a comprehensive analysis framework on generalisation opportunities of scientific and clinical impact, based on which new data-driven domain generalisation and adaptation methods can be stimulated.
CS$^2$: A Controllable and Simultaneous Synthesizer of Images and Annotations with Minimal Human Intervention
Jun 20, 2022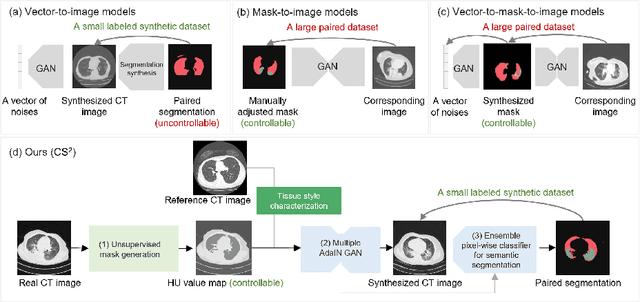
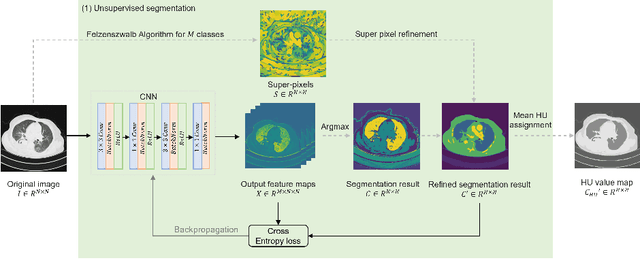
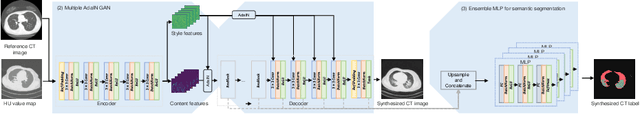
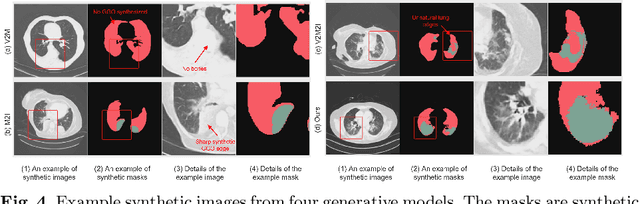
Abstract:The destitution of image data and corresponding expert annotations limit the training capacities of AI diagnostic models and potentially inhibit their performance. To address such a problem of data and label scarcity, generative models have been developed to augment the training datasets. Previously proposed generative models usually require manually adjusted annotations (e.g., segmentation masks) or need pre-labeling. However, studies have found that these pre-labeling based methods can induce hallucinating artifacts, which might mislead the downstream clinical tasks, while manual adjustment could be onerous and subjective. To avoid manual adjustment and pre-labeling, we propose a novel controllable and simultaneous synthesizer (dubbed CS$^2$) in this study to generate both realistic images and corresponding annotations at the same time. Our CS$^2$ model is trained and validated using high resolution CT (HRCT) data collected from COVID-19 patients to realize an efficient infections segmentation with minimal human intervention. Our contributions include 1) a conditional image synthesis network that receives both style information from reference CT images and structural information from unsupervised segmentation masks, and 2) a corresponding segmentation mask synthesis network to automatically segment these synthesized images simultaneously. Our experimental studies on HRCT scans collected from COVID-19 patients demonstrate that our CS$^2$ model can lead to realistic synthesized datasets and promising segmentation results of COVID infections compared to the state-of-the-art nnUNet trained and fine-tuned in a fully supervised manner.
Unsupervised Image Registration Towards Enhancing Performance and Explainability in Cardiac And Brain Image Analysis
Mar 07, 2022



Abstract:Magnetic Resonance Imaging (MRI) typically recruits multiple sequences (defined here as "modalities"). As each modality is designed to offer different anatomical and functional clinical information, there are evident disparities in the imaging content across modalities. Inter- and intra-modality affine and non-rigid image registration is an essential medical image analysis process in clinical imaging, as for example before imaging biomarkers need to be derived and clinically evaluated across different MRI modalities, time phases and slices. Although commonly needed in real clinical scenarios, affine and non-rigid image registration is not extensively investigated using a single unsupervised model architecture. In our work, we present an un-supervised deep learning registration methodology which can accurately model affine and non-rigid trans-formations, simultaneously. Moreover, inverse-consistency is a fundamental inter-modality registration property that is not considered in deep learning registration algorithms. To address inverse-consistency, our methodology performs bi-directional cross-modality image synthesis to learn modality-invariant latent rep-resentations, while involves two factorised transformation networks and an inverse-consistency loss to learn topology-preserving anatomical transformations. Overall, our model (named "FIRE") shows improved performances against the reference standard baseline method on multi-modality brain 2D and 3D MRI and intra-modality cardiac 4D MRI data experiments.
Robust Weakly Supervised Learning for COVID-19 Recognition Using Multi-Center CT Images
Dec 09, 2021



Abstract:The world is currently experiencing an ongoing pandemic of an infectious disease named coronavirus disease 2019 (i.e., COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Computed Tomography (CT) plays an important role in assessing the severity of the infection and can also be used to identify those symptomatic and asymptomatic COVID-19 carriers. With a surge of the cumulative number of COVID-19 patients, radiologists are increasingly stressed to examine the CT scans manually. Therefore, an automated 3D CT scan recognition tool is highly in demand since the manual analysis is time-consuming for radiologists and their fatigue can cause possible misjudgment. However, due to various technical specifications of CT scanners located in different hospitals, the appearance of CT images can be significantly different leading to the failure of many automated image recognition approaches. The multi-domain shift problem for the multi-center and multi-scanner studies is therefore nontrivial that is also crucial for a dependable recognition and critical for reproducible and objective diagnosis and prognosis. In this paper, we proposed a COVID-19 CT scan recognition model namely coronavirus information fusion and diagnosis network (CIFD-Net) that can efficiently handle the multi-domain shift problem via a new robust weakly supervised learning paradigm. Our model can resolve the problem of different appearance in CT scan images reliably and efficiently while attaining higher accuracy compared to other state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge