Haochuan Jiang
Mind the Gap: Promoting Missing Modality Brain Tumor Segmentation with Alignment
Sep 28, 2024Abstract:Brain tumor segmentation is often based on multiple magnetic resonance imaging (MRI). However, in clinical practice, certain modalities of MRI may be missing, which presents an even more difficult scenario. To cope with this challenge, knowledge distillation has emerged as one promising strategy. However, recent efforts typically overlook the modality gaps and thus fail to learn invariant feature representations across different modalities. Such drawback consequently leads to limited performance for both teachers and students. To ameliorate these problems, in this paper, we propose a novel paradigm that aligns latent features of involved modalities to a well-defined distribution anchor. As a major contribution, we prove that our novel training paradigm ensures a tight evidence lower bound, thus theoretically certifying its effectiveness. Extensive experiments on different backbones validate that the proposed paradigm can enable invariant feature representations and produce a teacher with narrowed modality gaps. This further offers superior guidance for missing modality students, achieving an average improvement of 1.75 on dice score.
MedMAP: Promoting Incomplete Multi-modal Brain Tumor Segmentation with Alignment
Aug 18, 2024Abstract:Brain tumor segmentation is often based on multiple magnetic resonance imaging (MRI). However, in clinical practice, certain modalities of MRI may be missing, which presents a more difficult scenario. To cope with this challenge, Knowledge Distillation, Domain Adaption, and Shared Latent Space have emerged as commonly promising strategies. However, recent efforts typically overlook the modality gaps and thus fail to learn important invariant feature representations across different modalities. Such drawback consequently leads to limited performance for missing modality models. To ameliorate these problems, pre-trained models are used in natural visual segmentation tasks to minimize the gaps. However, promising pre-trained models are often unavailable in medical image segmentation tasks. Along this line, in this paper, we propose a novel paradigm that aligns latent features of involved modalities to a well-defined distribution anchor as the substitution of the pre-trained model}. As a major contribution, we prove that our novel training paradigm ensures a tight evidence lower bound, thus theoretically certifying its effectiveness. Extensive experiments on different backbones validate that the proposed paradigm can enable invariant feature representations and produce models with narrowed modality gaps. Models with our alignment paradigm show their superior performance on both BraTS2018 and BraTS2020 datasets.
Generalized W-Net: Arbitrary-style Chinese Character Synthesization
Jun 10, 2024Abstract:Synthesizing Chinese characters with consistent style using few stylized examples is challenging. Existing models struggle to generate arbitrary style characters with limited examples. In this paper, we propose the Generalized W-Net, a novel class of W-shaped architectures that addresses this. By incorporating Adaptive Instance Normalization and introducing multi-content, our approach can synthesize Chinese characters in any desired style, even with limited examples. It handles seen and unseen styles during training and can generate new character contents. Experimental results demonstrate the effectiveness of our approach.
W-Net: One-Shot Arbitrary-Style Chinese Character Generation with Deep Neural Networks
Jun 10, 2024
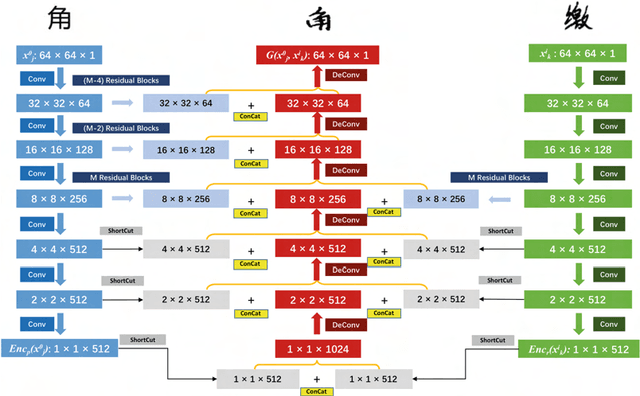


Abstract:Due to the huge category number, the sophisticated combinations of various strokes and radicals, and the free writing or printing styles, generating Chinese characters with diverse styles is always considered as a difficult task. In this paper, an efficient and generalized deep framework, namely, the W-Net, is introduced for the one-shot arbitrary-style Chinese character generation task. Specifically, given a single character (one-shot) with a specific style (e.g., a printed font or hand-writing style), the proposed W-Net model is capable of learning and generating any arbitrary characters sharing the style similar to the given single character. Such appealing property was rarely seen in the literature. We have compared the proposed W-Net framework to many other competitive methods. Experimental results showed the proposed method is significantly superior in the one-shot setting.
Rethinking Information Loss in Medical Image Segmentation with Various-sized Targets
Mar 28, 2024



Abstract:Medical image segmentation presents the challenge of segmenting various-size targets, demanding the model to effectively capture both local and global information. Despite recent efforts using CNNs and ViTs to predict annotations of different scales, these approaches often struggle to effectively balance the detection of targets across varying sizes. Simply utilizing local information from CNNs and global relationships from ViTs without considering potential significant divergence in latent feature distributions may result in substantial information loss. To address this issue, in this paper, we will introduce a novel Stagger Network (SNet) and argues that a well-designed fusion structure can mitigate the divergence in latent feature distributions between CNNs and ViTs, thereby reducing information loss. Specifically, to emphasize both global dependencies and local focus, we design a Parallel Module to bridge the semantic gap. Meanwhile, we propose the Stagger Module, trying to fuse the selected features that are more semantically similar. An Information Recovery Module is further adopted to recover complementary information back to the network. As a key contribution, we theoretically analyze that the proposed parallel and stagger strategies would lead to less information loss, thus certifying the SNet's rationale. Experimental results clearly proved that the proposed SNet excels comparisons with recent SOTAs in segmenting on the Synapse dataset where targets are in various sizes. Besides, it also demonstrates superiority on the ACDC and the MoNuSeg datasets where targets are with more consistent dimensions.
UGformer for Robust Left Atrium and Scar Segmentation Across Scanners
Oct 11, 2022
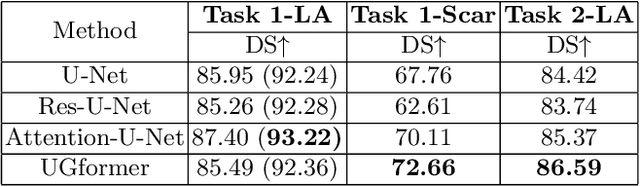
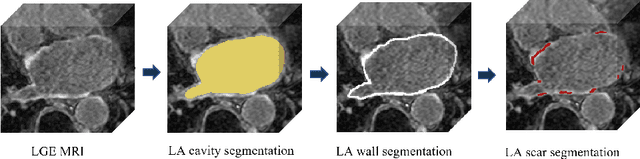
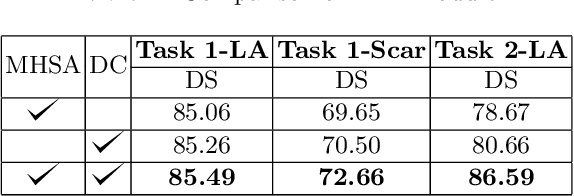
Abstract:Thanks to the capacity for long-range dependencies and robustness to irregular shapes, vision transformers and deformable convolutions are emerging as powerful vision techniques of segmentation.Meanwhile, Graph Convolution Networks (GCN) optimize local features based on global topological relationship modeling. Particularly, they have been proved to be effective in addressing issues in medical imaging segmentation tasks including multi-domain generalization for low-quality images. In this paper, we present a novel, effective, and robust framework for medical image segmentation, namely, UGformer. It unifies novel transformer blocks, GCN bridges, and convolution decoders originating from U-Net to predict left atriums (LAs) and LA scars. We have identified two appealing findings of the proposed UGformer: 1). an enhanced transformer module with deformable convolutions to improve the blending of the transformer information with convolutional information and help predict irregular LAs and scar shapes. 2). Using a bridge incorporating GCN to further overcome the difficulty of capturing condition inconsistency across different Magnetic Resonance Images scanners with various inconsistent domain information. The proposed UGformer model exhibits outstanding ability to segment the left atrium and scar on the LAScarQS 2022 dataset, outperforming several recent state-of-the-arts.
MyoPS: A Benchmark of Myocardial Pathology Segmentation Combining Three-Sequence Cardiac Magnetic Resonance Images
Jan 10, 2022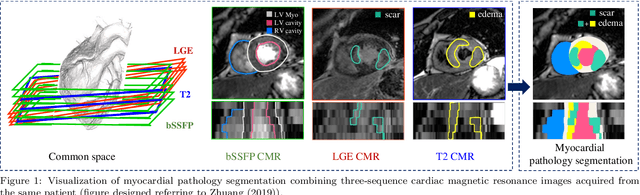
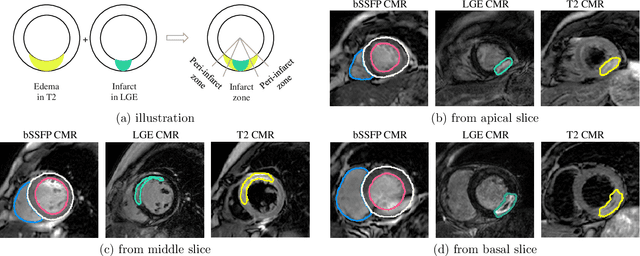
Abstract:Assessment of myocardial viability is essential in diagnosis and treatment management of patients suffering from myocardial infarction, and classification of pathology on myocardium is the key to this assessment. This work defines a new task of medical image analysis, i.e., to perform myocardial pathology segmentation (MyoPS) combining three-sequence cardiac magnetic resonance (CMR) images, which was first proposed in the MyoPS challenge, in conjunction with MICCAI 2020. The challenge provided 45 paired and pre-aligned CMR images, allowing algorithms to combine the complementary information from the three CMR sequences for pathology segmentation. In this article, we provide details of the challenge, survey the works from fifteen participants and interpret their methods according to five aspects, i.e., preprocessing, data augmentation, learning strategy, model architecture and post-processing. In addition, we analyze the results with respect to different factors, in order to examine the key obstacles and explore potential of solutions, as well as to provide a benchmark for future research. We conclude that while promising results have been reported, the research is still in the early stage, and more in-depth exploration is needed before a successful application to the clinics. Note that MyoPS data and evaluation tool continue to be publicly available upon registration via its homepage (www.sdspeople.fudan.edu.cn/zhuangxiahai/0/myops20/).
Max-Fusion U-Net for Multi-Modal Pathology Segmentation with Attention and Dynamic Resampling
Sep 05, 2020



Abstract:Automatic segmentation of multi-sequence (multi-modal) cardiac MR (CMR) images plays a significant role in diagnosis and management for a variety of cardiac diseases. However, the performance of relevant algorithms is significantly affected by the proper fusion of the multi-modal information. Furthermore, particular diseases, such as myocardial infarction, display irregular shapes on images and occupy small regions at random locations. These facts make pathology segmentation of multi-modal CMR images a challenging task. In this paper, we present the Max-Fusion U-Net that achieves improved pathology segmentation performance given aligned multi-modal images of LGE, T2-weighted, and bSSFP modalities. Specifically, modality-specific features are extracted by dedicated encoders. Then they are fused with the pixel-wise maximum operator. Together with the corresponding encoding features, these representations are propagated to decoding layers with U-Net skip-connections. Furthermore, a spatial-attention module is applied in the last decoding layer to encourage the network to focus on those small semantically meaningful pathological regions that trigger relatively high responses by the network neurons. We also use a simple image patch extraction strategy to dynamically resample training examples with varying spacial and batch sizes. With limited GPU memory, this strategy reduces the imbalance of classes and forces the model to focus on regions around the interested pathology. It further improves segmentation accuracy and reduces the mis-classification of pathology. We evaluate our methods using the Myocardial pathology segmentation (MyoPS) combining the multi-sequence CMR dataset which involves three modalities. Extensive experiments demonstrate the effectiveness of the proposed model which outperforms the related baselines.
* 13 pages, 7 figures, conference paper
Semi-supervised Pathology Segmentation with Disentangled Representations
Sep 05, 2020
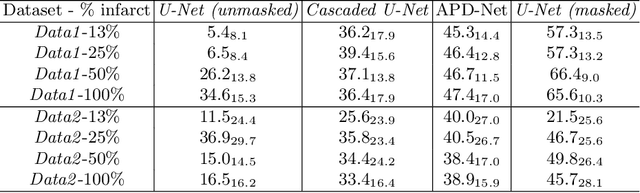
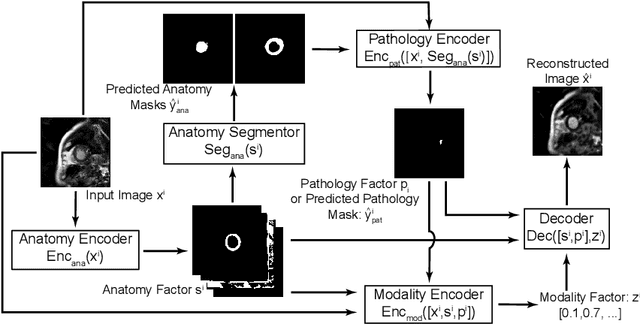
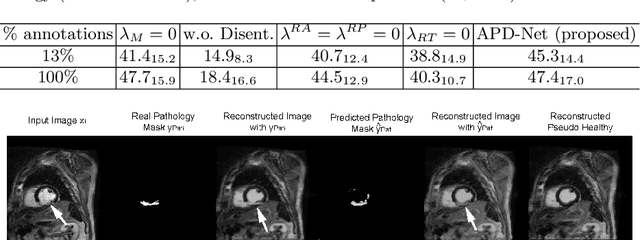
Abstract:Automated pathology segmentation remains a valuable diagnostic tool in clinical practice. However, collecting training data is challenging. Semi-supervised approaches by combining labelled and unlabelled data can offer a solution to data scarcity. An approach to semi-supervised learning relies on reconstruction objectives (as self-supervision objectives) that learns in a joint fashion suitable representations for the task. Here, we propose Anatomy-Pathology Disentanglement Network (APD-Net), a pathology segmentation model that attempts to learn jointly for the first time: disentanglement of anatomy, modality, and pathology. The model is trained in a semi-supervised fashion with new reconstruction losses directly aiming to improve pathology segmentation with limited annotations. In addition, a joint optimization strategy is proposed to fully take advantage of the available annotations. We evaluate our methods with two private cardiac infarction segmentation datasets with LGE-MRI scans. APD-Net can perform pathology segmentation with few annotations, maintain performance with different amounts of supervision, and outperform related deep learning methods.
* 12 Pages, 4 figures
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge