Scott Semple
Semi-supervised Pathology Segmentation with Disentangled Representations
Sep 05, 2020
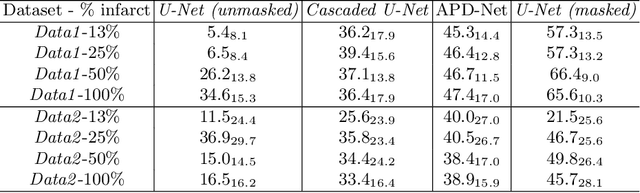
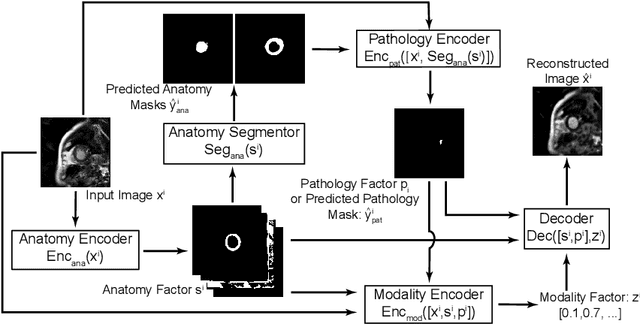
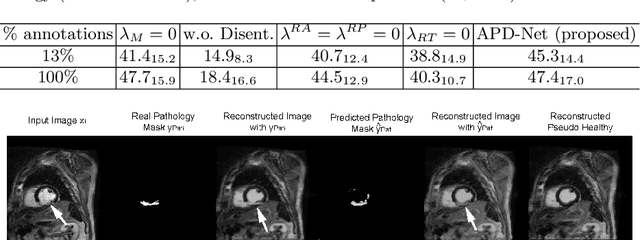
Abstract:Automated pathology segmentation remains a valuable diagnostic tool in clinical practice. However, collecting training data is challenging. Semi-supervised approaches by combining labelled and unlabelled data can offer a solution to data scarcity. An approach to semi-supervised learning relies on reconstruction objectives (as self-supervision objectives) that learns in a joint fashion suitable representations for the task. Here, we propose Anatomy-Pathology Disentanglement Network (APD-Net), a pathology segmentation model that attempts to learn jointly for the first time: disentanglement of anatomy, modality, and pathology. The model is trained in a semi-supervised fashion with new reconstruction losses directly aiming to improve pathology segmentation with limited annotations. In addition, a joint optimization strategy is proposed to fully take advantage of the available annotations. We evaluate our methods with two private cardiac infarction segmentation datasets with LGE-MRI scans. APD-Net can perform pathology segmentation with few annotations, maintain performance with different amounts of supervision, and outperform related deep learning methods.
* 12 Pages, 4 figures
Disentangle, align and fuse for multimodal and zero-shot image segmentation
Nov 12, 2019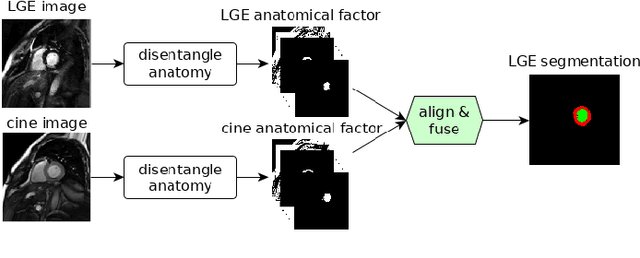
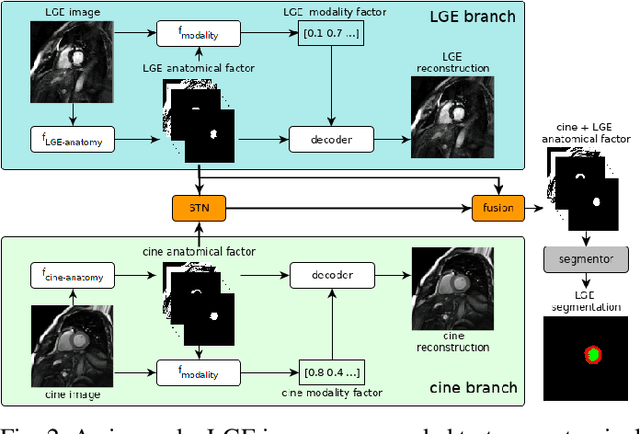
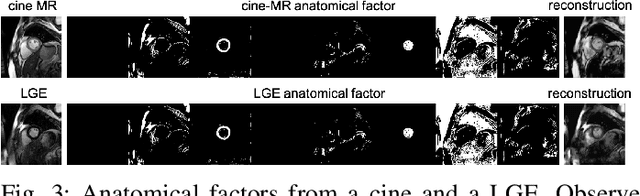
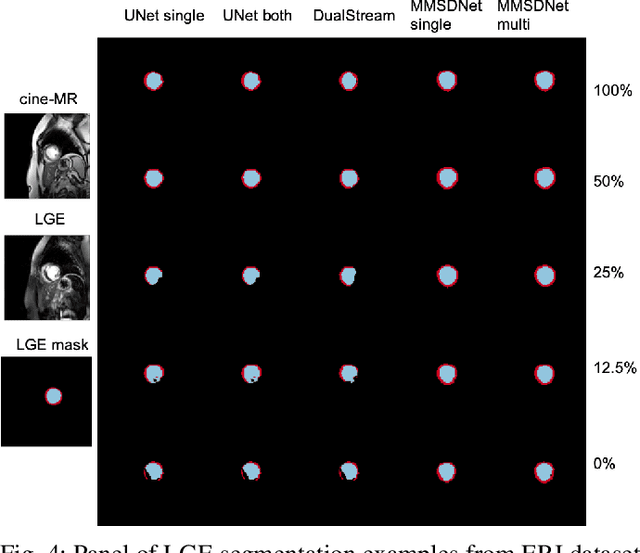
Abstract:Magnetic resonance (MR) protocols rely on several sequences to properly assess pathology and organ status. Yet, despite advances in image analysis we tend to treat each sequence, here termed modality, in isolation. Taking advantage of the information shared between modalities (largely an organ's anatomy) is beneficial for multi-modality multi-input processing and learning. However, we must overcome inherent anatomical misregistrations and disparities in signal intensity across the modalities to claim this benefit. We present a method that offers improved segmentation accuracy of the modality of interest (over a single input model), by learning to leverage information present in other modalities, enabling semi-supervised and zero shot learning. Core to our method is learning a disentangled decomposition into anatomical and imaging factors. Shared anatomical factors from the different inputs are jointly processed and fused to extract more accurate segmentation masks. Image misregistrations are corrected with a Spatial Transformer Network, that non-linearly aligns the anatomical factors. The imaging factor captures signal intensity characteristics across different modality data, and is used for image reconstruction, enabling semi-supervised learning. Temporal and slice pairing between inputs are learned dynamically. We demonstrate applications in Late Gadolinium Enhanced (LGE) and Blood Oxygenation Level Dependent (BOLD) cardiac segmentation, as well as in T2 abdominal segmentation.
Factorised spatial representation learning: application in semi-supervised myocardial segmentation
Nov 02, 2018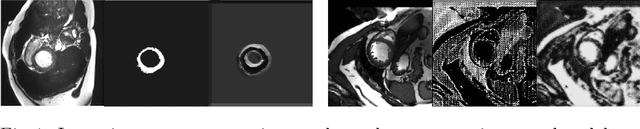
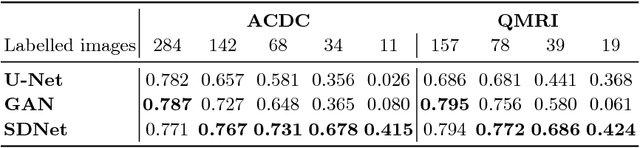
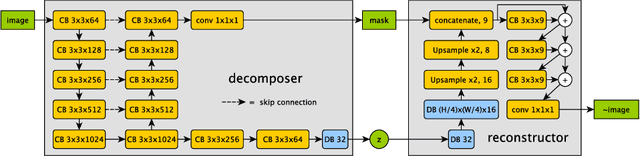
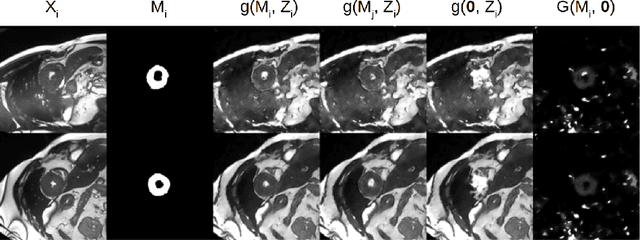
Abstract:The success and generalisation of deep learning algorithms heavily depend on learning good feature representations. In medical imaging this entails representing anatomical information, as well as properties related to the specific imaging setting. Anatomical information is required to perform further analysis, whereas imaging information is key to disentangle scanner variability and potential artefacts. The ability to factorise these would allow for training algorithms only on the relevant information according to the task. To date, such factorisation has not been attempted. In this paper, we propose a methodology of latent space factorisation relying on the cycle-consistency principle. As an example application, we consider cardiac MR segmentation, where we separate information related to the myocardium from other features related to imaging and surrounding substructures. We demonstrate the proposed method's utility in a semi-supervised setting: we use very few labelled images together with many unlabelled images to train a myocardium segmentation neural network. Specifically, we achieve comparable performance to fully supervised networks using a fraction of labelled images in experiments on ACDC and a dataset from Edinburgh Imaging Facility QMRI. Code will be made available at https://github.com/agis85/spatial_factorisation.
A Distance Oriented Kalman Filter Particle Swarm Optimizer Applied to Multi-Modality Image Registration
Mar 20, 2018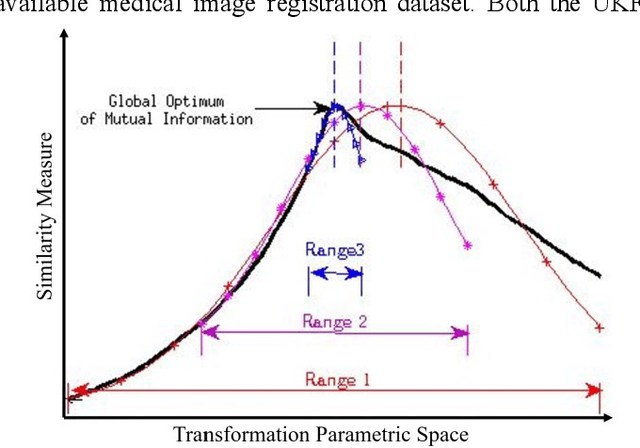
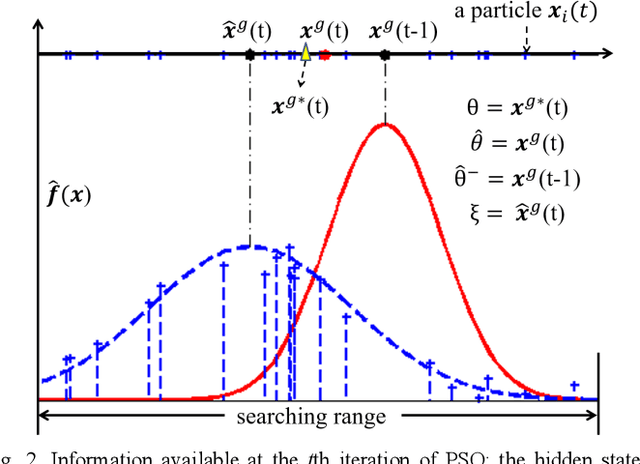
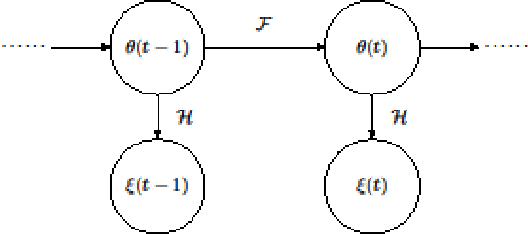
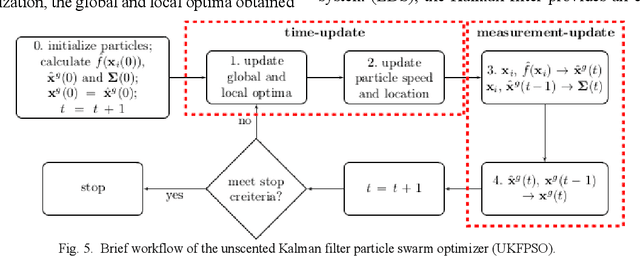
Abstract:In this paper we describe improvements to the particle swarm optimizer (PSO) made by inclusion of an unscented Kalman filter to guide particle motion. We demonstrate the effectiveness of the unscented Kalman filter PSO by comparing it with the original PSO algorithm and its variants designed to improve performance. The PSOs were tested firstly on a number of common synthetic benchmarking functions, and secondly applied to a practical three-dimensional image registration problem. The proposed methods displayed better performances for 4 out of 8 benchmark functions, and reduced the target registration errors by at least 2mm when registering down-sampled benchmark brain images. Our methods also demonstrated an ability to align images featuring motion related artefacts which all other methods failed to register. These new PSO methods provide a novel, efficient mechanism to integrate prior knowledge into each iteration of the optimization process, which can enhance the accuracy and speed of convergence in the application of medical image registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge