Sihan Wang
Decoupled Entity Representation Learning for Pinterest Ads Ranking
Sep 04, 2025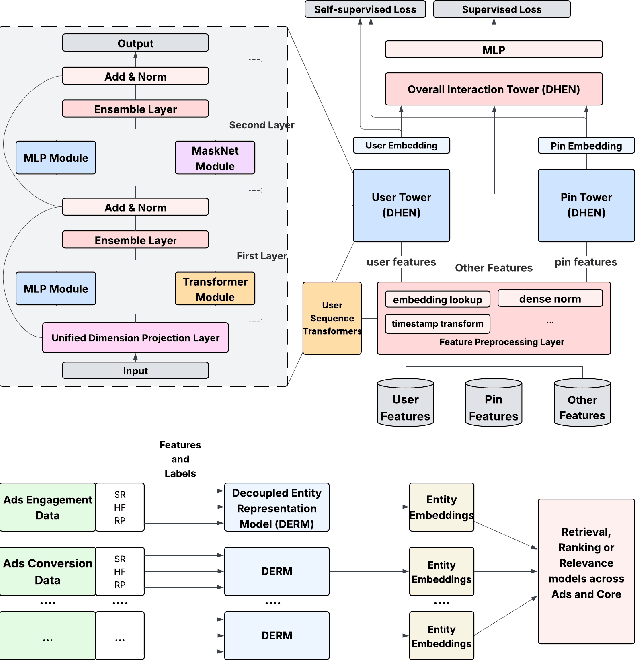



Abstract:In this paper, we introduce a novel framework following an upstream-downstream paradigm to construct user and item (Pin) embeddings from diverse data sources, which are essential for Pinterest to deliver personalized Pins and ads effectively. Our upstream models are trained on extensive data sources featuring varied signals, utilizing complex architectures to capture intricate relationships between users and Pins on Pinterest. To ensure scalability of the upstream models, entity embeddings are learned, and regularly refreshed, rather than real-time computation, allowing for asynchronous interaction between the upstream and downstream models. These embeddings are then integrated as input features in numerous downstream tasks, including ad retrieval and ranking models for CTR and CVR predictions. We demonstrate that our framework achieves notable performance improvements in both offline and online settings across various downstream tasks. This framework has been deployed in Pinterest's production ad ranking systems, resulting in significant gains in online metrics.
Decoupled Functional Evaluation of Autonomous Driving Models via Feature Map Quality Scoring
Aug 12, 2025Abstract:End-to-end models are emerging as the mainstream in autonomous driving perception and planning. However, the lack of explicit supervision signals for intermediate functional modules leads to opaque operational mechanisms and limited interpretability, making it challenging for traditional methods to independently evaluate and train these modules. Pioneering in the issue, this study builds upon the feature map-truth representation similarity-based evaluation framework and proposes an independent evaluation method based on Feature Map Convergence Score (FMCS). A Dual-Granularity Dynamic Weighted Scoring System (DG-DWSS) is constructed, formulating a unified quantitative metric - Feature Map Quality Score - to enable comprehensive evaluation of the quality of feature maps generated by functional modules. A CLIP-based Feature Map Quality Evaluation Network (CLIP-FMQE-Net) is further developed, combining feature-truth encoders and quality score prediction heads to enable real-time quality analysis of feature maps generated by functional modules. Experimental results on the NuScenes dataset demonstrate that integrating our evaluation module into the training improves 3D object detection performance, achieving a 3.89 percent gain in NDS. These results verify the effectiveness of our method in enhancing feature representation quality and overall model performance.
CineMyoPS: Segmenting Myocardial Pathologies from Cine Cardiac MR
Jul 03, 2025



Abstract:Myocardial infarction (MI) is a leading cause of death worldwide. Late gadolinium enhancement (LGE) and T2-weighted cardiac magnetic resonance (CMR) imaging can respectively identify scarring and edema areas, both of which are essential for MI risk stratification and prognosis assessment. Although combining complementary information from multi-sequence CMR is useful, acquiring these sequences can be time-consuming and prohibitive, e.g., due to the administration of contrast agents. Cine CMR is a rapid and contrast-free imaging technique that can visualize both motion and structural abnormalities of the myocardium induced by acute MI. Therefore, we present a new end-to-end deep neural network, referred to as CineMyoPS, to segment myocardial pathologies, \ie scars and edema, solely from cine CMR images. Specifically, CineMyoPS extracts both motion and anatomy features associated with MI. Given the interdependence between these features, we design a consistency loss (resembling the co-training strategy) to facilitate their joint learning. Furthermore, we propose a time-series aggregation strategy to integrate MI-related features across the cardiac cycle, thereby enhancing segmentation accuracy for myocardial pathologies. Experimental results on a multi-center dataset demonstrate that CineMyoPS achieves promising performance in myocardial pathology segmentation, motion estimation, and anatomy segmentation.
Learning Concept-Driven Logical Rules for Interpretable and Generalizable Medical Image Classification
May 20, 2025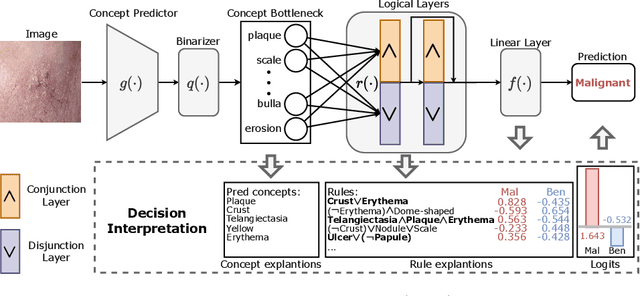
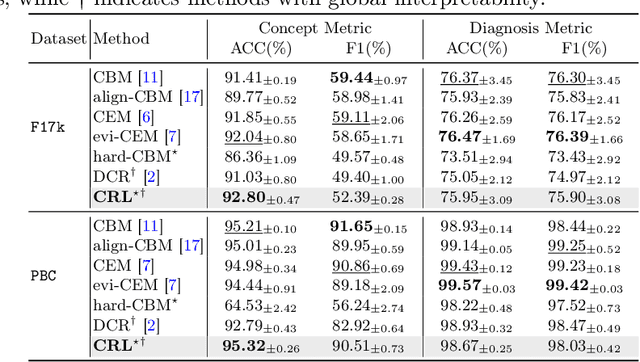
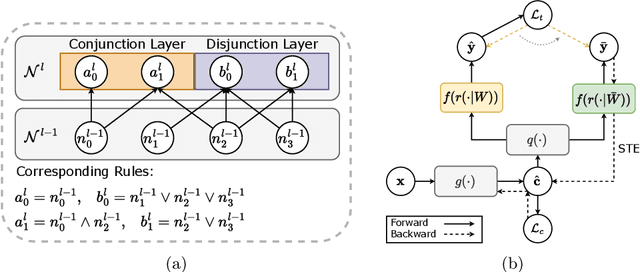
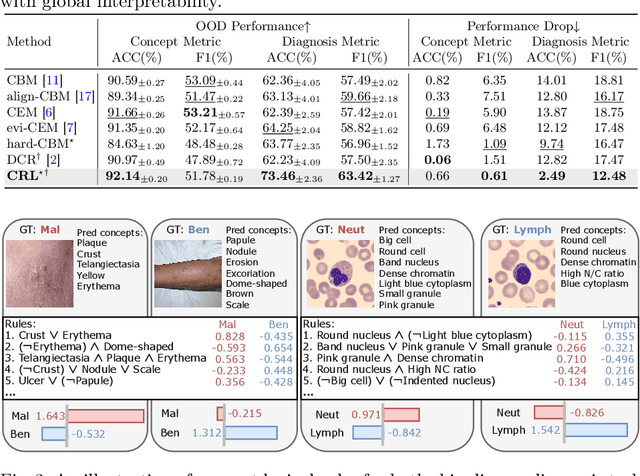
Abstract:The pursuit of decision safety in clinical applications highlights the potential of concept-based methods in medical imaging. While these models offer active interpretability, they often suffer from concept leakages, where unintended information within soft concept representations undermines both interpretability and generalizability. Moreover, most concept-based models focus solely on local explanations (instance-level), neglecting the global decision logic (dataset-level). To address these limitations, we propose Concept Rule Learner (CRL), a novel framework to learn Boolean logical rules from binarized visual concepts. CRL employs logical layers to capture concept correlations and extract clinically meaningful rules, thereby providing both local and global interpretability. Experiments on two medical image classification tasks show that CRL achieves competitive performance with existing methods while significantly improving generalizability to out-of-distribution data. The code of our work is available at https://github.com/obiyoag/crl.
Empowering Medical Multi-Agents with Clinical Consultation Flow for Dynamic Diagnosis
Mar 19, 2025



Abstract:Traditional AI-based healthcare systems often rely on single-modal data, limiting diagnostic accuracy due to incomplete information. However, recent advancements in foundation models show promising potential for enhancing diagnosis combining multi-modal information. While these models excel in static tasks, they struggle with dynamic diagnosis, failing to manage multi-turn interactions and often making premature diagnostic decisions due to insufficient persistence in information collection.To address this, we propose a multi-agent framework inspired by consultation flow and reinforcement learning (RL) to simulate the entire consultation process, integrating multiple clinical information for effective diagnosis. Our approach incorporates a hierarchical action set, structured from clinic consultation flow and medical textbook, to effectively guide the decision-making process. This strategy improves agent interactions, enabling them to adapt and optimize actions based on the dynamic state. We evaluated our framework on a public dynamic diagnosis benchmark. The proposed framework evidentially improves the baseline methods and achieves state-of-the-art performance compared to existing foundation model-based methods.
InDeed: Interpretable image deep decomposition with guaranteed generalizability
Jan 02, 2025



Abstract:Image decomposition aims to analyze an image into elementary components, which is essential for numerous downstream tasks and also by nature provides certain interpretability to the analysis. Deep learning can be powerful for such tasks, but surprisingly their combination with a focus on interpretability and generalizability is rarely explored. In this work, we introduce a novel framework for interpretable deep image decomposition, combining hierarchical Bayesian modeling and deep learning to create an architecture-modularized and model-generalizable deep neural network (DNN). The proposed framework includes three steps: (1) hierarchical Bayesian modeling of image decomposition, (2) transforming the inference problem into optimization tasks, and (3) deep inference via a modularized Bayesian DNN. We further establish a theoretical connection between the loss function and the generalization error bound, which inspires a new test-time adaptation approach for out-of-distribution scenarios. We instantiated the application using two downstream tasks, \textit{i.e.}, image denoising, and unsupervised anomaly detection, and the results demonstrated improved generalizability as well as interpretability of our methods. The source code will be released upon the acceptance of this paper.
Prototyping and Experimental Results for Environment-Aware Millimeter Wave Beam Alignment via Channel Knowledge Map
Mar 13, 2024Abstract:Channel knowledge map (CKM), which aims to directly reflect the intrinsic channel properties of the local wireless environment, is a novel technique for achieving environmentaware communication. In this paper, to alleviate the large training overhead in millimeter wave (mmWave) beam alignment, an environment-aware and training-free beam alignment prototype is established based on a typical CKM, termed beam index map (BIM). To this end, a general CKM construction method is first presented, and an indoor BIM is constructed offline to learn the candidate transmit and receive beam index pairs for each grid in the experimental area. Furthermore, based on the location information of the receiver (or the dynamic obstacles) from the ultra-wide band (UWB) positioning system, the established BIM is used to achieve training-free beam alignment by directly providing the beam indexes for the transmitter and receiver. Three typical scenarios are considered in the experiment, including quasi-static environment with line-of-sight (LoS) link, quasistatic environment without LoS link and dynamic environment. Besides, the receiver orientation measured from the gyroscope is also used to help CKM predict more accurate beam indexes. The experiment results show that compared with the benchmark location-based beam alignment strategy, the CKM-based beam alignment strategy can achieve much higher received power, which is close to that achieved by exhaustive beam search, but with significantly reduced training overhead.
Aligning Multi-Sequence CMR Towards Fully Automated Myocardial Pathology Segmentation
Feb 07, 2023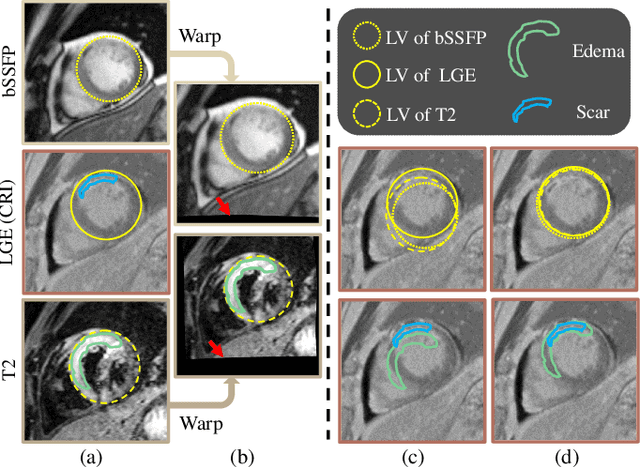
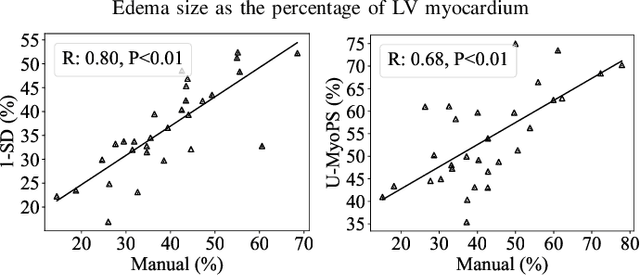
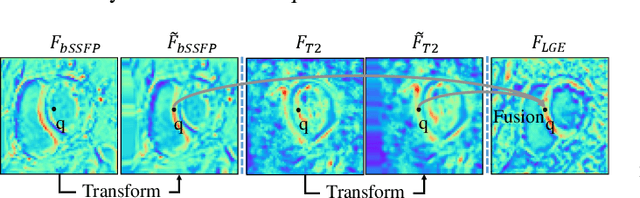
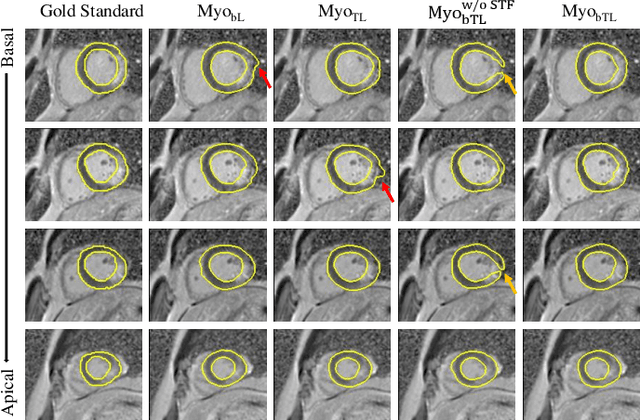
Abstract:Myocardial pathology segmentation (MyoPS) is critical for the risk stratification and treatment planning of myocardial infarction (MI). Multi-sequence cardiac magnetic resonance (MS-CMR) images can provide valuable information. For instance, balanced steady-state free precession cine sequences present clear anatomical boundaries, while late gadolinium enhancement and T2-weighted CMR sequences visualize myocardial scar and edema of MI, respectively. Existing methods usually fuse anatomical and pathological information from different CMR sequences for MyoPS, but assume that these images have been spatially aligned. However, MS-CMR images are usually unaligned due to the respiratory motions in clinical practices, which poses additional challenges for MyoPS. This work presents an automatic MyoPS framework for unaligned MS-CMR images. Specifically, we design a combined computing model for simultaneous image registration and information fusion, which aggregates multi-sequence features into a common space to extract anatomical structures (i.e., myocardium). Consequently, we can highlight the informative regions in the common space via the extracted myocardium to improve MyoPS performance, considering the spatial relationship between myocardial pathologies and myocardium. Experiments on a private MS-CMR dataset and a public dataset from the MYOPS2020 challenge show that our framework could achieve promising performance for fully automatic MyoPS.
Unsupervised Cardiac Segmentation Utilizing Synthesized Images from Anatomical Labels
Jan 15, 2023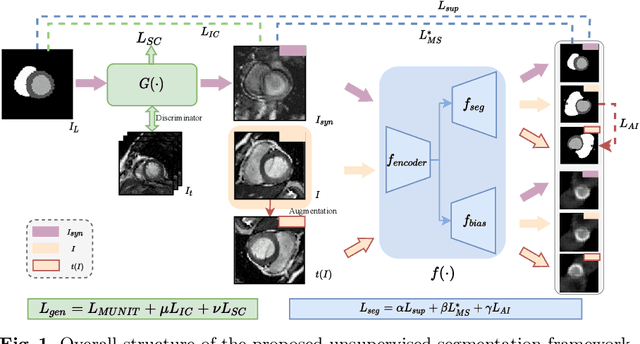
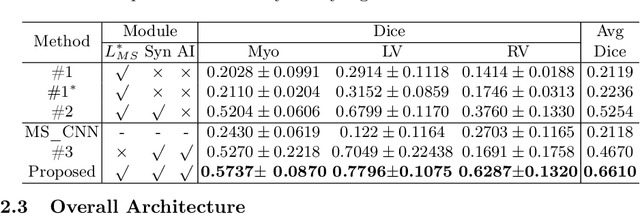
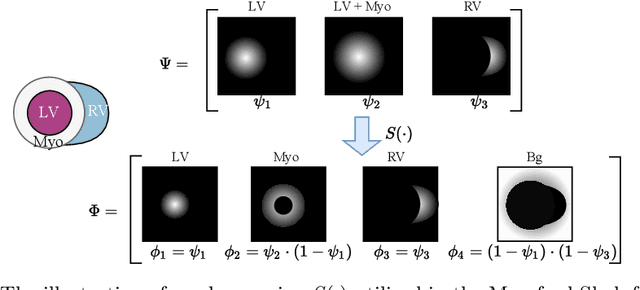
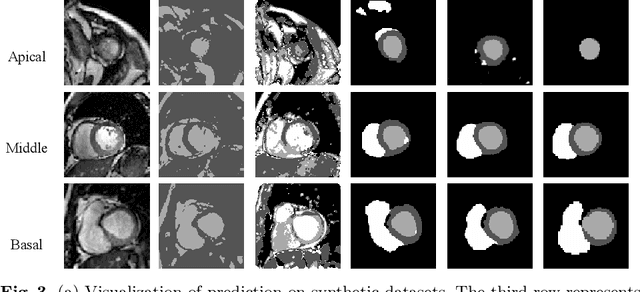
Abstract:Cardiac segmentation is in great demand for clinical practice. Due to the enormous labor of manual delineation, unsupervised segmentation is desired. The ill-posed optimization problem of this task is inherently challenging, requiring well-designed constraints. In this work, we propose an unsupervised framework for multi-class segmentation with both intensity and shape constraints. Firstly, we extend a conventional non-convex energy function as an intensity constraint and implement it with U-Net. For shape constraint, synthetic images are generated from anatomical labels via image-to-image translation, as shape supervision for the segmentation network. Moreover, augmentation invariance is applied to facilitate the segmentation network to learn the latent features in terms of shape. We evaluated the proposed framework using the public datasets from MICCAI2019 MSCMR Challenge and achieved promising results on cardiac MRIs with Dice scores of 0.5737, 0.7796, and 0.6287 in Myo, LV, and RV, respectively.
MyoPS-Net: Myocardial Pathology Segmentation with Flexible Combination of Multi-Sequence CMR Images
Nov 06, 2022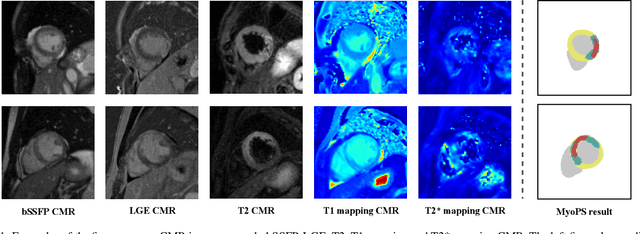



Abstract:Myocardial pathology segmentation (MyoPS) can be a prerequisite for the accurate diagnosis and treatment planning of myocardial infarction. However, achieving this segmentation is challenging, mainly due to the inadequate and indistinct information from an image. In this work, we develop an end-to-end deep neural network, referred to as MyoPS-Net, to flexibly combine five-sequence cardiac magnetic resonance (CMR) images for MyoPS. To extract precise and adequate information, we design an effective yet flexible architecture to extract and fuse cross-modal features. This architecture can tackle different numbers of CMR images and complex combinations of modalities, with output branches targeting specific pathologies. To impose anatomical knowledge on the segmentation results, we first propose a module to regularize myocardium consistency and localize the pathologies, and then introduce an inclusiveness loss to utilize relations between myocardial scars and edema. We evaluated the proposed MyoPS-Net on two datasets, i.e., a private one consisting of 50 paired multi-sequence CMR images and a public one from MICCAI2020 MyoPS Challenge. Experimental results showed that MyoPS-Net could achieve state-of-the-art performance in various scenarios. Note that in practical clinics, the subjects may not have full sequences, such as missing LGE CMR or mapping CMR scans. We therefore conducted extensive experiments to investigate the performance of the proposed method in dealing with such complex combinations of different CMR sequences. Results proved the superiority and generalizability of MyoPS-Net, and more importantly, indicated a practical clinical application.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge