Yibo Gao
S3-CoT: Self-Sampled Succinct Reasoning Enables Efficient Chain-of-Thought LLMs
Feb 02, 2026Abstract:Large language models (LLMs) equipped with chain-of-thought (CoT) achieve strong performance and offer a window into LLM behavior. However, recent evidence suggests that improvements in CoT capabilities often come with redundant reasoning processes, motivating a key question: Can LLMs acquire a fast-thinking mode analogous to human System 1 reasoning? To explore this, our study presents a self-sampling framework based on activation steering for efficient CoT learning. Our method can induce style-aligned and variable-length reasoning traces from target LLMs themselves without any teacher guidance, thereby alleviating a central bottleneck of SFT-based methods-the scarcity of high-quality supervision data. Using filtered data by gold answers, we perform SFT for efficient CoT learning with (i) a human-like dual-cognitive system, and (ii) a progressive compression curriculum. Furthermore, we explore a self-evolution regime in which SFT is driven solely by prediction-consistent data of variable-length variants, eliminating the need for gold answers. Extensive experiments on math benchmarks, together with cross-domain generalization tests in medicine, show that our method yields stable improvements for both general and R1-style LLMs. Our data and model checkpoints can be found at https://github.com/DYR1/S3-CoT.
From Latent Signals to Reflection Behavior: Tracing Meta-Cognitive Activation Trajectory in R1-Style LLMs
Feb 02, 2026Abstract:R1-style LLMs have attracted growing attention for their capacity for self-reflection, yet the internal mechanisms underlying such behavior remain unclear. To bridge this gap, we anchor on the onset of reflection behavior and trace its layer-wise activation trajectory. Using the logit lens to read out token-level semantics, we uncover a structured progression: (i) Latent-control layers, where an approximate linear direction encodes the semantics of thinking budget; (ii) Semantic-pivot layers, where discourse-level cues, including turning-point and summarization cues, surface and dominate the probability mass; and (iii) Behavior-overt layers, where the likelihood of reflection-behavior tokens begins to rise until they become highly likely to be sampled. Moreover, our targeted interventions uncover a causal chain across these stages: prompt-level semantics modulate the projection of activations along latent-control directions, thereby inducing competition between turning-point and summarization cues in semantic-pivot layers, which in turn regulates the sampling likelihood of reflection-behavior tokens in behavior-overt layers. Collectively, our findings suggest a human-like meta-cognitive process-progressing from latent monitoring, to discourse-level regulation, and to finally overt self-reflection. Our analysis code can be found at https://github.com/DYR1/S3-CoT.
Learning Concept-Driven Logical Rules for Interpretable and Generalizable Medical Image Classification
May 20, 2025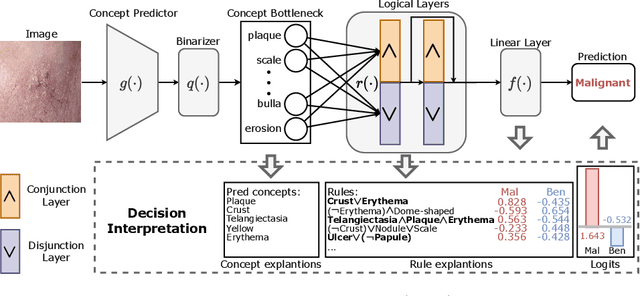
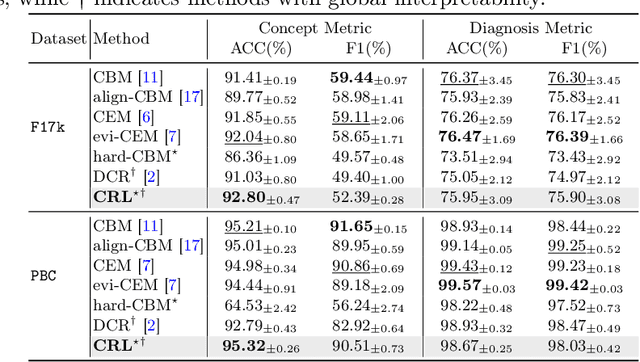
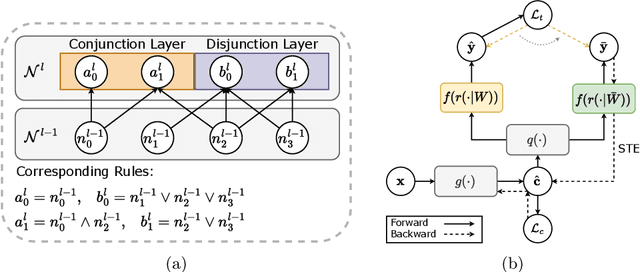
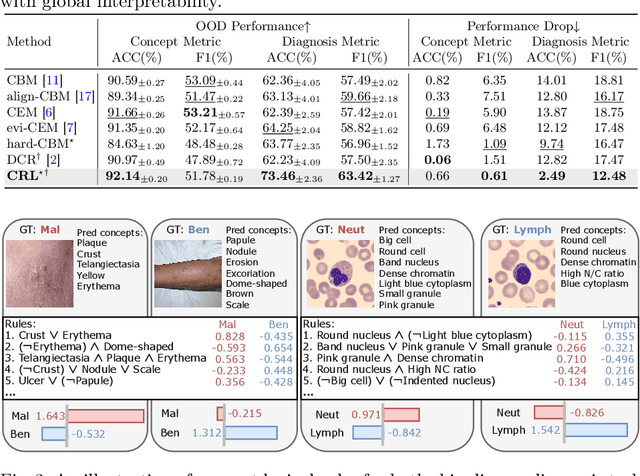
Abstract:The pursuit of decision safety in clinical applications highlights the potential of concept-based methods in medical imaging. While these models offer active interpretability, they often suffer from concept leakages, where unintended information within soft concept representations undermines both interpretability and generalizability. Moreover, most concept-based models focus solely on local explanations (instance-level), neglecting the global decision logic (dataset-level). To address these limitations, we propose Concept Rule Learner (CRL), a novel framework to learn Boolean logical rules from binarized visual concepts. CRL employs logical layers to capture concept correlations and extract clinically meaningful rules, thereby providing both local and global interpretability. Experiments on two medical image classification tasks show that CRL achieves competitive performance with existing methods while significantly improving generalizability to out-of-distribution data. The code of our work is available at https://github.com/obiyoag/crl.
Empowering Medical Multi-Agents with Clinical Consultation Flow for Dynamic Diagnosis
Mar 19, 2025



Abstract:Traditional AI-based healthcare systems often rely on single-modal data, limiting diagnostic accuracy due to incomplete information. However, recent advancements in foundation models show promising potential for enhancing diagnosis combining multi-modal information. While these models excel in static tasks, they struggle with dynamic diagnosis, failing to manage multi-turn interactions and often making premature diagnostic decisions due to insufficient persistence in information collection.To address this, we propose a multi-agent framework inspired by consultation flow and reinforcement learning (RL) to simulate the entire consultation process, integrating multiple clinical information for effective diagnosis. Our approach incorporates a hierarchical action set, structured from clinic consultation flow and medical textbook, to effectively guide the decision-making process. This strategy improves agent interactions, enabling them to adapt and optimize actions based on the dynamic state. We evaluated our framework on a public dynamic diagnosis benchmark. The proposed framework evidentially improves the baseline methods and achieves state-of-the-art performance compared to existing foundation model-based methods.
Evidential Concept Embedding Models: Towards Reliable Concept Explanations for Skin Disease Diagnosis
Jun 27, 2024Abstract:Due to the high stakes in medical decision-making, there is a compelling demand for interpretable deep learning methods in medical image analysis. Concept Bottleneck Models (CBM) have emerged as an active interpretable framework incorporating human-interpretable concepts into decision-making. However, their concept predictions may lack reliability when applied to clinical diagnosis, impeding concept explanations' quality. To address this, we propose an evidential Concept Embedding Model (evi-CEM), which employs evidential learning to model the concept uncertainty. Additionally, we offer to leverage the concept uncertainty to rectify concept misalignments that arise when training CBMs using vision-language models without complete concept supervision. With the proposed methods, we can enhance concept explanations' reliability for both supervised and label-efficient settings. Furthermore, we introduce concept uncertainty for effective test-time intervention. Our evaluation demonstrates that evi-CEM achieves superior performance in terms of concept prediction, and the proposed concept rectification effectively mitigates concept misalignments for label-efficient training. Our code is available at https://github.com/obiyoag/evi-CEM.
Multi-Center Fetal Brain Tissue Annotation (FeTA) Challenge 2022 Results
Feb 08, 2024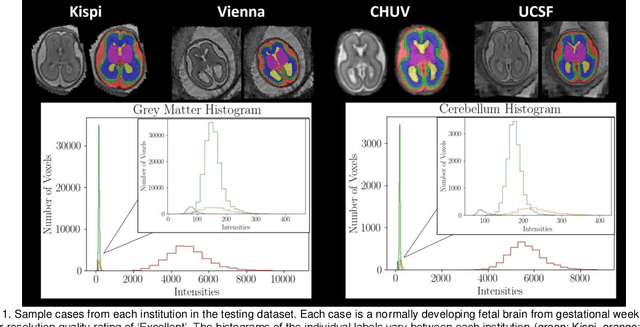
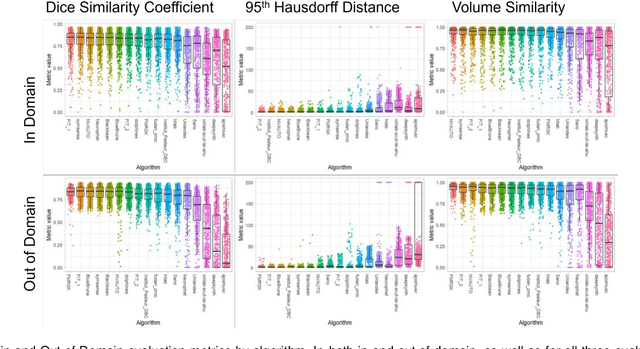
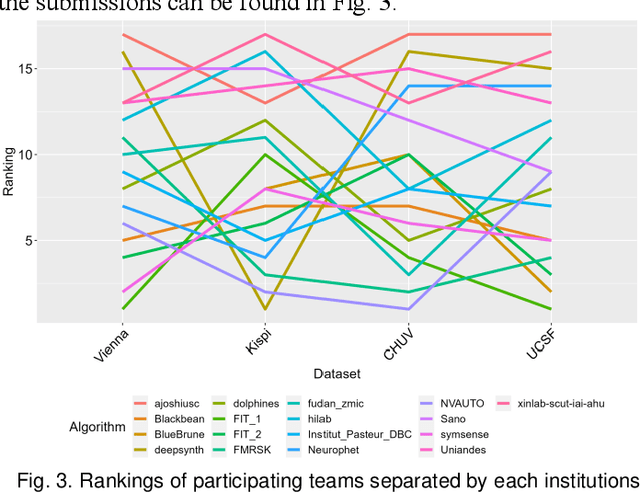
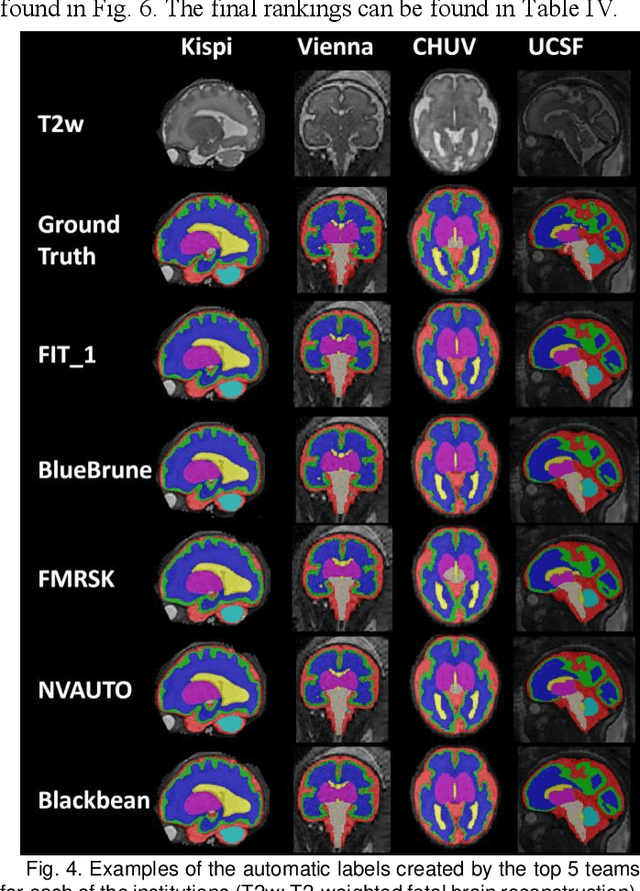
Abstract:Segmentation is a critical step in analyzing the developing human fetal brain. There have been vast improvements in automatic segmentation methods in the past several years, and the Fetal Brain Tissue Annotation (FeTA) Challenge 2021 helped to establish an excellent standard of fetal brain segmentation. However, FeTA 2021 was a single center study, and the generalizability of algorithms across different imaging centers remains unsolved, limiting real-world clinical applicability. The multi-center FeTA Challenge 2022 focuses on advancing the generalizability of fetal brain segmentation algorithms for magnetic resonance imaging (MRI). In FeTA 2022, the training dataset contained images and corresponding manually annotated multi-class labels from two imaging centers, and the testing data contained images from these two imaging centers as well as two additional unseen centers. The data from different centers varied in many aspects, including scanners used, imaging parameters, and fetal brain super-resolution algorithms applied. 16 teams participated in the challenge, and 17 algorithms were evaluated. Here, a detailed overview and analysis of the challenge results are provided, focusing on the generalizability of the submissions. Both in- and out of domain, the white matter and ventricles were segmented with the highest accuracy, while the most challenging structure remains the cerebral cortex due to anatomical complexity. The FeTA Challenge 2022 was able to successfully evaluate and advance generalizability of multi-class fetal brain tissue segmentation algorithms for MRI and it continues to benchmark new algorithms. The resulting new methods contribute to improving the analysis of brain development in utero.
BayeSeg: Bayesian Modeling for Medical Image Segmentation with Interpretable Generalizability
Mar 03, 2023Abstract:Due to the cross-domain distribution shift aroused from diverse medical imaging systems, many deep learning segmentation methods fail to perform well on unseen data, which limits their real-world applicability. Recent works have shown the benefits of extracting domain-invariant representations on domain generalization. However, the interpretability of domain-invariant features remains a great challenge. To address this problem, we propose an interpretable Bayesian framework (BayeSeg) through Bayesian modeling of image and label statistics to enhance model generalizability for medical image segmentation. Specifically, we first decompose an image into a spatial-correlated variable and a spatial-variant variable, assigning hierarchical Bayesian priors to explicitly force them to model the domain-stable shape and domain-specific appearance information respectively. Then, we model the segmentation as a locally smooth variable only related to the shape. Finally, we develop a variational Bayesian framework to infer the posterior distributions of these explainable variables. The framework is implemented with neural networks, and thus is referred to as deep Bayesian segmentation. Quantitative and qualitative experimental results on prostate segmentation and cardiac segmentation tasks have shown the effectiveness of our proposed method. Moreover, we investigated the interpretability of BayeSeg by explaining the posteriors and analyzed certain factors that affect the generalization ability through further ablation studies. Our code will be released via https://zmiclab.github.io/projects.html, once the manuscript is accepted for publication.
Joint Modeling of Image and Label Statistics for Enhancing Model Generalizability of Medical Image Segmentation
Jun 09, 2022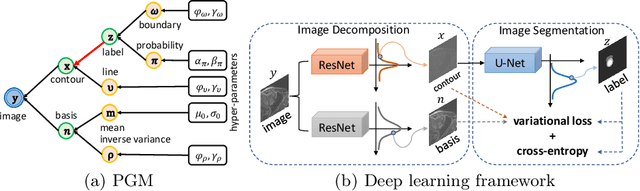
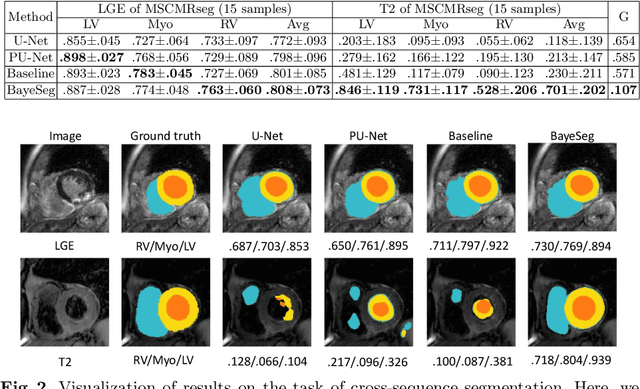
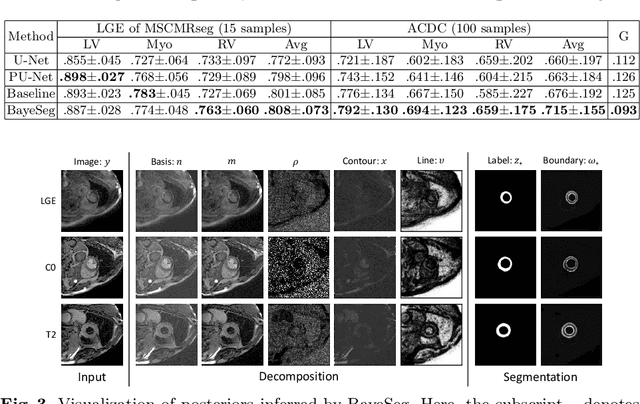
Abstract:Although supervised deep-learning has achieved promising performance in medical image segmentation, many methods cannot generalize well on unseen data, limiting their real-world applicability. To address this problem, we propose a deep learning-based Bayesian framework, which jointly models image and label statistics, utilizing the domain-irrelevant contour of a medical image for segmentation. Specifically, we first decompose an image into components of contour and basis. Then, we model the expected label as a variable only related to the contour. Finally, we develop a variational Bayesian framework to infer the posterior distributions of these variables, including the contour, the basis, and the label. The framework is implemented with neural networks, thus is referred to as deep Bayesian segmentation. Results on the task of cross-sequence cardiac MRI segmentation show that our method set a new state of the art for model generalizability. Particularly, the BayeSeg model trained with LGE MRI generalized well on T2 images and outperformed other models with great margins, i.e., over 0.47 in terms of average Dice. Our code is available at https://zmiclab.github.io/projects.html.
Towards Self-supervised and Weight-preserving Neural Architecture Search
Jun 08, 2022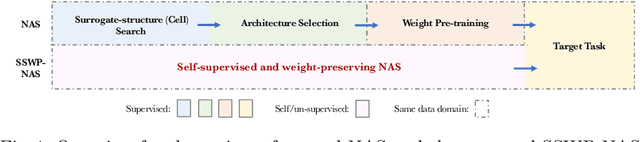
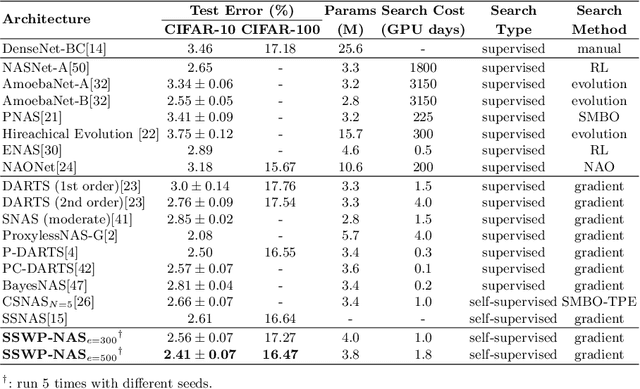
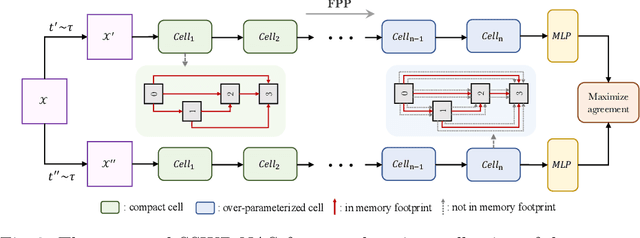
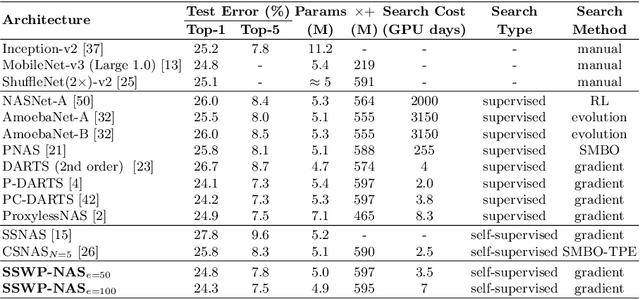
Abstract:Neural architecture search (NAS) algorithms save tremendous labor from human experts. Recent advancements further reduce the computational overhead to an affordable level. However, it is still cumbersome to deploy the NAS techniques in real-world applications due to the fussy procedures and the supervised learning paradigm. In this work, we propose the self-supervised and weight-preserving neural architecture search (SSWP-NAS) as an extension of the current NAS framework by allowing the self-supervision and retaining the concomitant weights discovered during the search stage. As such, we simplify the workflow of NAS to a one-stage and proxy-free procedure. Experiments show that the architectures searched by the proposed framework achieve state-of-the-art accuracy on CIFAR-10, CIFAR-100, and ImageNet datasets without using manual labels. Moreover, we show that employing the concomitant weights as initialization consistently outperforms the random initialization and the two-stage weight pre-training method by a clear margin under semi-supervised learning scenarios. Codes are publicly available at https://github.com/LzVv123456/SSWP-NAS.
Plantbot: A New ROS-based Robot Platform for Fast Building and Developing
Apr 08, 2018
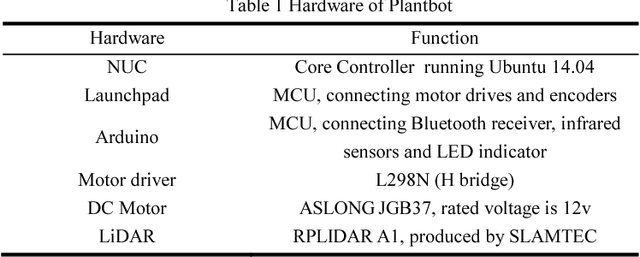

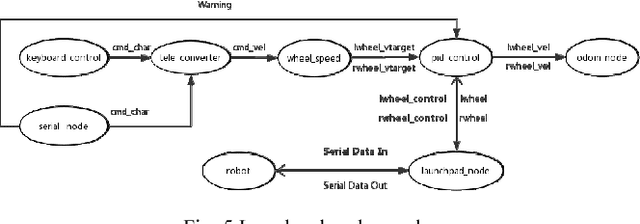
Abstract:In recent years, the Robot Operating System (ROS) is developing rapidly and has been widely used in robotics research because of its flexible, open source, and extensive advantages. In scientific research, the corresponding hardware platform is indispensable for the experiment. In the field of mobile robots, PR2, Turtlebot2, and Fetch are commonly used as research platforms. Although these platforms are fully functional and widely used, they are expensive and bulky. What\u2019s more, these robots are not easily redesigned and expanded according to requirements. To overcome these limitations, we propose Plantbot, an easy-building, low-cost robot platform that can be redesigned and expanded according to requirements. It can be applied to not only fast algorithm verification, but also simple factory inspection and ROS teaching. This article describes this robot platform from several aspects such as hardware design, kinematics, and control methods. At last two experiments, SLAM and Navigation, on the robot platform are performed. The source code of this platform is available.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge