Xin Gao
Computer Science Program, Computer, Electrical and Mathematical Sciences and Engineering Division, King Abdullah University of Science and Technology, Center of Excellence for Smart Health, Center of Excellence on Generative AI, King Abdullah University of Science and Technology
MedTri: A Platform for Structured Medical Report Normalization to Enhance Vision-Language Pretraining
Feb 25, 2026Abstract:Medical vision-language pretraining increasingly relies on medical reports as large-scale supervisory signals; however, raw reports often exhibit substantial stylistic heterogeneity, variable length, and a considerable amount of image-irrelevant content. Although text normalization is frequently adopted as a preprocessing step in prior work, its design principles and empirical impact on vision-language pretraining remain insufficiently and systematically examined. In this study, we present MedTri, a deployable normalization framework for medical vision-language pretraining that converts free-text reports into a unified [Anatomical Entity: Radiologic Description + Diagnosis Category] triplet. This structured, anatomy-grounded normalization preserves essential morphological and spatial information while removing stylistic noise and image-irrelevant content, providing consistent and image-grounded textual supervision at scale. Across multiple datasets spanning both X-ray and computed tomography (CT) modalities, we demonstrate that structured, anatomy-grounded text normalization is an important factor in medical vision-language pretraining quality, yielding consistent improvements over raw reports and existing normalization baselines. In addition, we illustrate how this normalization can easily support modular text-level augmentation strategies, including knowledge enrichment and anatomy-grounded counterfactual supervision, which provide complementary gains in robustness and generalization without altering the core normalization process. Together, our results position structured text normalization as a critical and generalizable preprocessing component for medical vision-language learning, while MedTri provides this normalization platform. Code and data will be released at https://github.com/Arturia-Pendragon-Iris/MedTri.
ChartVerse: Scaling Chart Reasoning via Reliable Programmatic Synthesis from Scratch
Jan 20, 2026Abstract:Chart reasoning is a critical capability for Vision Language Models (VLMs). However, the development of open-source models is severely hindered by the lack of high-quality training data. Existing datasets suffer from a dual challenge: synthetic charts are often simplistic and repetitive, while the associated QA pairs are prone to hallucinations and lack the reasoning depth required for complex tasks. To bridge this gap, we propose ChartVerse, a scalable framework designed to synthesize complex charts and reliable reasoning data from scratch. (1) To address the bottleneck of simple patterns, we first introduce Rollout Posterior Entropy (RPE), a novel metric that quantifies chart complexity. Guided by RPE, we develop complexity-aware chart coder to autonomously synthesize diverse, high-complexity charts via executable programs. (2) To guarantee reasoning rigor, we develop truth-anchored inverse QA synthesis. Diverging from standard generation, we adopt an answer-first paradigm: we extract deterministic answers directly from the source code, generate questions conditional on these anchors, and enforce strict consistency verification. To further elevate difficulty and reasoning depth, we filter samples based on model fail-rate and distill high-quality Chain-of-Thought (CoT) reasoning. We curate ChartVerse-SFT-600K and ChartVerse-RL-40K using Qwen3-VL-30B-A3B-Thinking as the teacher. Experimental results demonstrate that ChartVerse-8B achieves state-of-the-art performance, notably surpassing its teacher and rivaling the stronger Qwen3-VL-32B-Thinking.
Scientific Image Synthesis: Benchmarking, Methodologies, and Downstream Utility
Jan 17, 2026Abstract:While synthetic data has proven effective for improving scientific reasoning in the text domain, multimodal reasoning remains constrained by the difficulty of synthesizing scientifically rigorous images. Existing Text-to-Image (T2I) models often produce outputs that are visually plausible yet scientifically incorrect, resulting in a persistent visual-logic divergence that limits their value for downstream reasoning. Motivated by recent advances in next-generation T2I models, we conduct a systematic study of scientific image synthesis across generation paradigms, evaluation, and downstream use. We analyze both direct pixel-based generation and programmatic synthesis, and propose ImgCoder, a logic-driven framework that follows an explicit "understand - plan - code" workflow to improve structural precision. To rigorously assess scientific correctness, we introduce SciGenBench, which evaluates generated images based on information utility and logical validity. Our evaluation reveals systematic failure modes in pixel-based models and highlights a fundamental expressiveness-precision trade-off. Finally, we show that fine-tuning Large Multimodal Models (LMMs) on rigorously verified synthetic scientific images yields consistent reasoning gains, with potential scaling trends analogous to the text domain, validating high-fidelity scientific synthesis as a viable path to unlocking massive multimodal reasoning capabilities.
Bridging Global Intent with Local Details: A Hierarchical Representation Approach for Semantic Validation in Text-to-SQL
Dec 28, 2025Abstract:Text-to-SQL translates natural language questions into SQL statements grounded in a target database schema. Ensuring the reliability and executability of such systems requires validating generated SQL, but most existing approaches focus only on syntactic correctness, with few addressing semantic validation (detecting misalignments between questions and SQL). As a consequence, effective semantic validation still faces two key challenges: capturing both global user intent and SQL structural details, and constructing high-quality fine-grained sub-SQL annotations. To tackle these, we introduce HEROSQL, a hierarchical SQL representation approach that integrates global intent (via Logical Plans, LPs) and local details (via Abstract Syntax Trees, ASTs). To enable better information propagation, we employ a Nested Message Passing Neural Network (NMPNN) to capture inherent relational information in SQL and aggregate schema-guided semantics across LPs and ASTs. Additionally, to generate high-quality negative samples, we propose an AST-driven sub-SQL augmentation strategy, supporting robust optimization of fine-grained semantic inconsistencies. Extensive experiments conducted on Text-to-SQL validation benchmarks (both in-domain and out-of-domain settings) demonstrate that our approach outperforms existing state-of-the-art methods, achieving an average 9.40% improvement of AUPRC and 12.35% of AUROC in identifying semantic inconsistencies. It excels at detecting fine-grained semantic errors, provides large language models with more granular feedback, and ultimately enhances the reliability and interpretability of data querying platforms.
GaussianEM: Model compositional and conformational heterogeneity using 3D Gaussians
Dec 25, 2025Abstract:Understanding protein flexibility and its dynamic interactions with other molecules is essential for protein function study. Cryogenic electron microscopy (cryo-EM) provides an opportunity to directly observe macromolecular dynamics. However, analyzing datasets that contain both continuous motions and discrete states remains highly challenging. Here we present GaussianEM, a Gaussian pseudo-atomic framework that simultaneously models compositional and conformational heterogeneity from experimental cryo-EM images. GaussianEM employs a two-encoder-one-decoder architecture to map an image to its individual Gaussian components, and represent structural variability through changes in Gaussian parameters. This approach provides an intuitive and interpretable description of conformational changes, preserves local structural consistency along the transition trajectories, and naturally bridges the gap between density-based models and corresponding atomic models. We demonstrate the effectiveness of GaussianEM on both simulated and experimental datasets.
Multi-View MRI Approach for Classification of MGMT Methylation in Glioblastoma Patients
Dec 16, 2025Abstract:The presence of MGMT promoter methylation significantly affects how well chemotherapy works for patients with Glioblastoma Multiforme (GBM). Currently, confirmation of MGMT promoter methylation relies on invasive brain tumor tissue biopsies. In this study, we explore radiogenomics techniques, a promising approach in precision medicine, to identify genetic markers from medical images. Using MRI scans and deep learning models, we propose a new multi-view approach that considers spatial relationships between MRI views to detect MGMT methylation status. Importantly, our method extracts information from all three views without using a complicated 3D deep learning model, avoiding issues associated with high parameter count, slow convergence, and substantial memory demands. We also introduce a new technique for tumor slice extraction and show its superiority over existing methods based on multiple evaluation metrics. By comparing our approach to state-of-the-art models, we demonstrate the efficacy of our method. Furthermore, we share a reproducible pipeline of published models, encouraging transparency and the development of robust diagnostic tools. Our study highlights the potential of non-invasive methods for identifying MGMT promoter methylation and contributes to advancing precision medicine in GBM treatment.
OpenDataArena: A Fair and Open Arena for Benchmarking Post-Training Dataset Value
Dec 16, 2025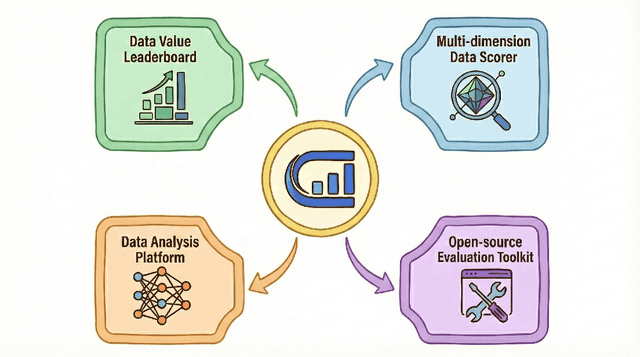
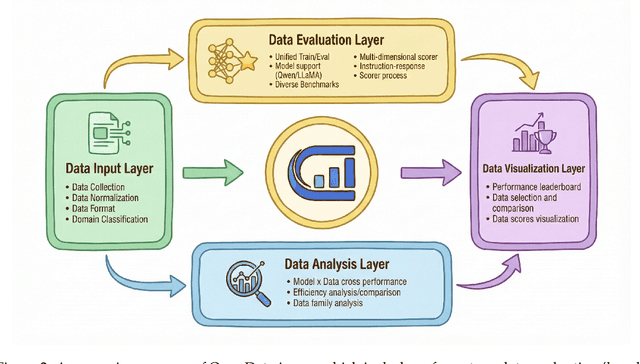
Abstract:The rapid evolution of Large Language Models (LLMs) is predicated on the quality and diversity of post-training datasets. However, a critical dichotomy persists: while models are rigorously benchmarked, the data fueling them remains a black box--characterized by opaque composition, uncertain provenance, and a lack of systematic evaluation. This opacity hinders reproducibility and obscures the causal link between data characteristics and model behaviors. To bridge this gap, we introduce OpenDataArena (ODA), a holistic and open platform designed to benchmark the intrinsic value of post-training data. ODA establishes a comprehensive ecosystem comprising four key pillars: (i) a unified training-evaluation pipeline that ensures fair, open comparisons across diverse models (e.g., Llama, Qwen) and domains; (ii) a multi-dimensional scoring framework that profiles data quality along tens of distinct axes; (iii) an interactive data lineage explorer to visualize dataset genealogy and dissect component sources; and (iv) a fully open-source toolkit for training, evaluation, and scoring to foster data research. Extensive experiments on ODA--covering over 120 training datasets across multiple domains on 22 benchmarks, validated by more than 600 training runs and 40 million processed data points--reveal non-trivial insights. Our analysis uncovers the inherent trade-offs between data complexity and task performance, identifies redundancy in popular benchmarks through lineage tracing, and maps the genealogical relationships across datasets. We release all results, tools, and configurations to democratize access to high-quality data evaluation. Rather than merely expanding a leaderboard, ODA envisions a shift from trial-and-error data curation to a principled science of Data-Centric AI, paving the way for rigorous studies on data mixing laws and the strategic composition of foundation models.
A data-physics hybrid generative model for patient-specific post-stroke motor rehabilitation using wearable sensor data
Dec 16, 2025Abstract:Dynamic prediction of locomotor capacity after stroke is crucial for tailoring rehabilitation, yet current assessments provide only static impairment scores and do not indicate whether patients can safely perform specific tasks such as slope walking or stair climbing. Here, we develop a data-physics hybrid generative framework that reconstructs an individual stroke survivor's neuromuscular control from a single 20 m level-ground walking trial and predicts task-conditioned locomotion across rehabilitation scenarios. The system combines wearable-sensor kinematics, a proportional-derivative physics controller, a population Healthy Motion Atlas, and goal-conditioned deep reinforcement learning with behaviour cloning and generative adversarial imitation learning to generate physically plausible, patient-specific gait simulations for slopes and stairs. In 11 stroke survivors, the personalized controllers preserved idiosyncratic gait patterns while improving joint-angle and endpoint fidelity by 4.73% and 12.10%, respectively, and reducing training time to 25.56% relative to a physics-only baseline. In a multicentre pilot involving 21 inpatients, clinicians who used our locomotion predictions to guide task selection and difficulty obtained larger gains in Fugl-Meyer lower-extremity scores over 28 days of standard rehabilitation than control clinicians (mean change 6.0 versus 3.7 points). These findings indicate that our generative, task-predictive framework can augment clinical decision-making in post-stroke gait rehabilitation and provide a template for dynamically personalized motor recovery strategies.
SafeRBench: A Comprehensive Benchmark for Safety Assessment in Large Reasoning Models
Nov 19, 2025Abstract:Large Reasoning Models (LRMs) improve answer quality through explicit chain-of-thought, yet this very capability introduces new safety risks: harmful content can be subtly injected, surface gradually, or be justified by misleading rationales within the reasoning trace. Existing safety evaluations, however, primarily focus on output-level judgments and rarely capture these dynamic risks along the reasoning process. In this paper, we present SafeRBench, the first benchmark that assesses LRM safety end-to-end -- from inputs and intermediate reasoning to final outputs. (1) Input Characterization: We pioneer the incorporation of risk categories and levels into input design, explicitly accounting for affected groups and severity, and thereby establish a balanced prompt suite reflecting diverse harm gradients. (2) Fine-Grained Output Analysis: We introduce a micro-thought chunking mechanism to segment long reasoning traces into semantically coherent units, enabling fine-grained evaluation across ten safety dimensions. (3) Human Safety Alignment: We validate LLM-based evaluations against human annotations specifically designed to capture safety judgments. Evaluations on 19 LRMs demonstrate that SafeRBench enables detailed, multidimensional safety assessment, offering insights into risks and protective mechanisms from multiple perspectives.
Revisiting Data Scaling Law for Medical Segmentation
Nov 17, 2025Abstract:The population loss of trained deep neural networks often exhibits power law scaling with the size of the training dataset, guiding significant performance advancements in deep learning applications. In this study, we focus on the scaling relationship with data size in the context of medical anatomical segmentation, a domain that remains underexplored. We analyze scaling laws for anatomical segmentation across 15 semantic tasks and 4 imaging modalities, demonstrating that larger datasets significantly improve segmentation performance, following similar scaling trends. Motivated by the topological isomorphism in images sharing anatomical structures, we evaluate the impact of deformation-guided augmentation strategies on data scaling laws, specifically random elastic deformation and registration-guided deformation. We also propose a novel, scalable image augmentation approach that generates diffeomorphic mappings from geodesic subspace based on image registration to introduce realistic deformation. Our experimental results demonstrate that both registered and generated deformation-based augmentation considerably enhance data utilization efficiency. The proposed generated deformation method notably achieves superior performance and accelerated convergence, surpassing standard power law scaling trends without requiring additional data. Overall, this work provides insights into the understanding of segmentation scalability and topological variation impact in medical imaging, thereby leading to more efficient model development with reduced annotation and computational costs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge