Changchun Yang
Xinhua Hospital Affiliated to Shanghai Jiao Tong University School of Medicine, Computer Science Program, Computer, Electrical and Mathematical Sciences and Engineering Division, King Abdullah University of Science and Technology, Center of Excellence for Smart Health, Center of Excellence on Generative AI, King Abdullah University of Science and Technology
PAST: A multimodal single-cell foundation model for histopathology and spatial transcriptomics in cancer
Jul 08, 2025Abstract:While pathology foundation models have transformed cancer image analysis, they often lack integration with molecular data at single-cell resolution, limiting their utility for precision oncology. Here, we present PAST, a pan-cancer single-cell foundation model trained on 20 million paired histopathology images and single-cell transcriptomes spanning multiple tumor types and tissue contexts. By jointly encoding cellular morphology and gene expression, PAST learns unified cross-modal representations that capture both spatial and molecular heterogeneity at the cellular level. This approach enables accurate prediction of single-cell gene expression, virtual molecular staining, and multimodal survival analysis directly from routine pathology slides. Across diverse cancers and downstream tasks, PAST consistently exceeds the performance of existing approaches, demonstrating robust generalizability and scalability. Our work establishes a new paradigm for pathology foundation models, providing a versatile tool for high-resolution spatial omics, mechanistic discovery, and precision cancer research.
An Inclusive Foundation Model for Generalizable Cytogenetics in Precision Oncology
May 21, 2025Abstract:Chromosome analysis is vital for diagnosing genetic disorders and guiding cancer therapy decisions through the identification of somatic clonal aberrations. However, developing an AI model are hindered by the overwhelming complexity and diversity of chromosomal abnormalities, requiring extensive annotation efforts, while automated methods remain task-specific and lack generalizability due to the scarcity of comprehensive datasets spanning diverse resource conditions. Here, we introduce CHROMA, a foundation model for cytogenomics, designed to overcome these challenges by learning generalizable representations of chromosomal abnormalities. Pre-trained on over 84,000 specimens (~4 million chromosomal images) via self-supervised learning, CHROMA outperforms other methods across all types of abnormalities, even when trained on fewer labelled data and more imbalanced datasets. By facilitating comprehensive mapping of instability and clonal leisons across various aberration types, CHROMA offers a scalable and generalizable solution for reliable and automated clinical analysis, reducing the annotation workload for experts and advancing precision oncology through the early detection of rare genomic abnormalities, enabling broad clinical AI applications and making advanced genomic analysis more accessible.
Improving Representation of High-frequency Components for Medical Foundation Models
Jul 26, 2024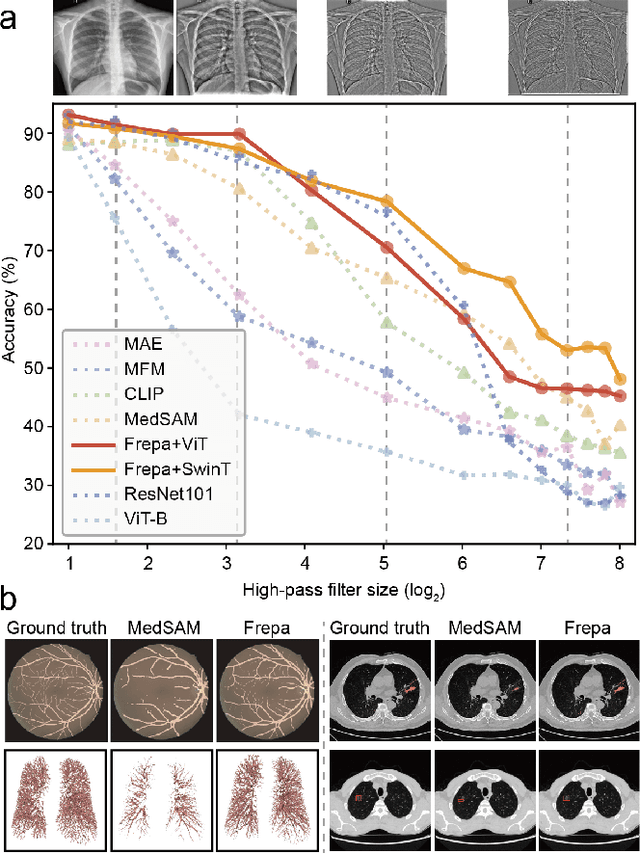

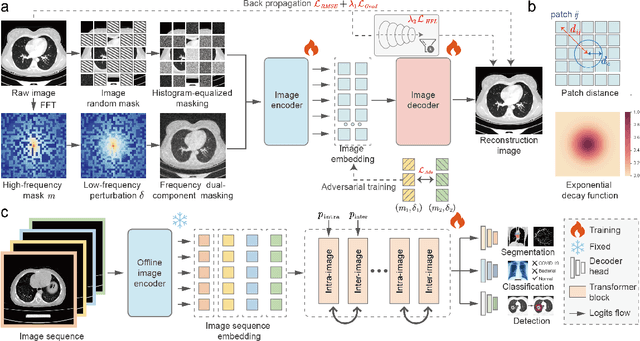

Abstract:Foundation models have recently attracted significant attention for their impressive generalizability across diverse downstream tasks. However, these models are demonstrated to exhibit great limitations in representing high-frequency components and fine-grained details. In many medical imaging tasks, the precise representation of such information is crucial due to the inherently intricate anatomical structures, sub-visual features, and complex boundaries involved. Consequently, the limited representation of prevalent foundation models can result in significant performance degradation or even failure in these tasks. To address these challenges, we propose a novel pretraining strategy, named Frequency-advanced Representation Autoencoder (Frepa). Through high-frequency masking and low-frequency perturbation combined with adversarial learning, Frepa encourages the encoder to effectively represent and preserve high-frequency components in the image embeddings. Additionally, we introduce an innovative histogram-equalized image masking strategy, extending the Masked Autoencoder approach beyond ViT to other architectures such as Swin Transformer and convolutional networks. We develop Frepa across nine medical modalities and validate it on 32 downstream tasks for both 2D images and 3D volume data. Without fine-tuning, Frepa can outperform other self-supervised pretraining methods and, in some cases, even surpasses task-specific trained models. This improvement is particularly significant for tasks involving fine-grained details, such as achieving up to a +15% increase in DSC for retina vessel segmentation and a +7% increase in IoU for lung nodule detection. Further experiments quantitatively reveal that Frepa enables superior high-frequency representations and preservation in the embeddings, underscoring its potential for developing more generalized and universal medical image foundation models.
Deep learning-driven pulmonary arteries and veins segmentation reveals demography-associated pulmonary vasculature anatomy
Apr 11, 2024Abstract:Pulmonary artery-vein segmentation is crucial for diagnosing pulmonary diseases and surgical planning, and is traditionally achieved by Computed Tomography Pulmonary Angiography (CTPA). However, concerns regarding adverse health effects from contrast agents used in CTPA have constrained its clinical utility. In contrast, identifying arteries and veins using non-contrast CT, a conventional and low-cost clinical examination routine, has long been considered impossible. Here we propose a High-abundant Pulmonary Artery-vein Segmentation (HiPaS) framework achieving accurate artery-vein segmentation on both non-contrast CT and CTPA across various spatial resolutions. HiPaS first performs spatial normalization on raw CT scans via a super-resolution module, and then iteratively achieves segmentation results at different branch levels by utilizing the low-level vessel segmentation as a prior for high-level vessel segmentation. We trained and validated HiPaS on our established multi-centric dataset comprising 1,073 CT volumes with meticulous manual annotation. Both quantitative experiments and clinical evaluation demonstrated the superior performance of HiPaS, achieving a dice score of 91.8% and a sensitivity of 98.0%. Further experiments demonstrated the non-inferiority of HiPaS segmentation on non-contrast CT compared to segmentation on CTPA. Employing HiPaS, we have conducted an anatomical study of pulmonary vasculature on 10,613 participants in China (five sites), discovering a new association between pulmonary vessel abundance and sex and age: vessel abundance is significantly higher in females than in males, and slightly decreases with age, under the controlling of lung volumes (p < 0.0001). HiPaS realizing accurate artery-vein segmentation delineates a promising avenue for clinical diagnosis and understanding pulmonary physiology in a non-invasive manner.
Efficient Bayesian Uncertainty Estimation for nnU-Net
Dec 12, 2022Abstract:The self-configuring nnU-Net has achieved leading performance in a large range of medical image segmentation challenges. It is widely considered as the model of choice and a strong baseline for medical image segmentation. However, despite its extraordinary performance, nnU-Net does not supply a measure of uncertainty to indicate its possible failure. This can be problematic for large-scale image segmentation applications, where data are heterogeneous and nnU-Net may fail without notice. In this work, we introduce a novel method to estimate nnU-Net uncertainty for medical image segmentation. We propose a highly effective scheme for posterior sampling of weight space for Bayesian uncertainty estimation. Different from previous baseline methods such as Monte Carlo Dropout and mean-field Bayesian Neural Networks, our proposed method does not require a variational architecture and keeps the original nnU-Net architecture intact, thereby preserving its excellent performance and ease of use. Additionally, we boost the segmentation performance over the original nnU-Net via marginalizing multi-modal posterior models. We applied our method on the public ACDC and M&M datasets of cardiac MRI and demonstrated improved uncertainty estimation over a range of baseline methods. The proposed method further strengthens nnU-Net for medical image segmentation in terms of both segmentation accuracy and quality control.
Photoacoustic digital brain: numerical modelling and image reconstruction via deep learning
Sep 19, 2021



Abstract:Photoacoustic tomography (PAT) is a newly developed medical imaging modality, which combines the advantages of pure optical imaging and ultrasound imaging, owning both high optical contrast and deep penetration depth. Very recently, PAT is studied in human brain imaging. Nevertheless, while ultrasound waves are passing through the human skull tissues, the strong acoustic attenuation and aberration will happen, which causes photoacoustic signals' distortion. In this work, we use 10 magnetic resonance angiography (MRA) human brain volumes, and manually segment them to obtain the 2D human brain numerical phantoms for PAT. The numerical phantoms contain six kinds of tissues which are scalp, skull, white matter, gray matter, blood vessel and cerebral cortex. For every numerical phantom, optical properties are assigned to every kind of tissues. Then, Monte-Carlo based optical simulation is deployed to obtain the photoacoustic initial pressure. Then, we made two k-wave simulation cases: one takes inhomogeneous medium and uneven sound velocity into consideration, and the other not. Then we use the sensor data of the former one as the input of U-net, and the sensor data of the latter one as the output of U-net to train the network. We randomly choose 7 human brain PA sinograms as the training dataset and 3 human brain PA sinograms as the testing set. The testing result shows that our method could correct the skull acoustic aberration and obtain the blood vessel distribution inside the human brain satisfactorily.
Compressed Sensing for Photoacoustic Computed Tomography Using an Untrained Neural Network
May 29, 2021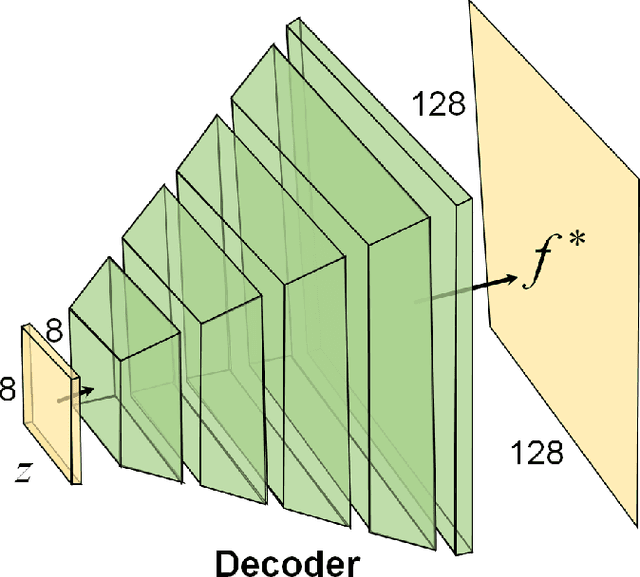
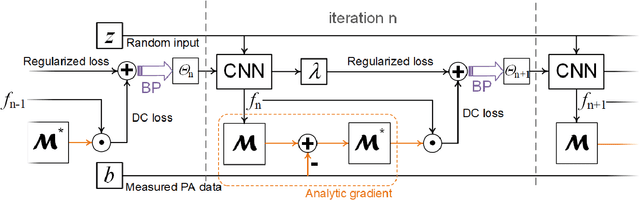
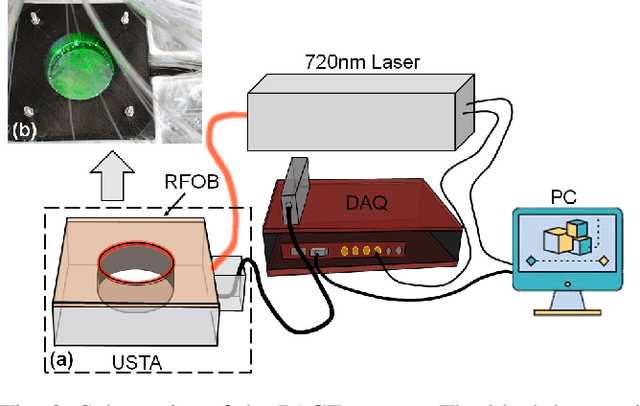
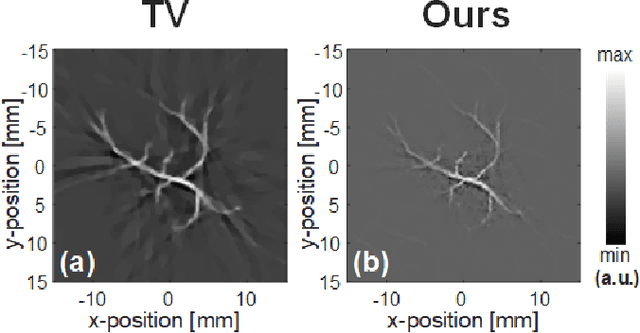
Abstract:Photoacoustic (PA) computed tomography (PACT) shows great potentials in various preclinical and clinical applications. A great number of measurements are the premise that obtains a high-quality image, which implies a low imaging rate or a high system cost. The artifacts or sidelobes could pollute the image if we decrease the number of measured channels or limit the detected view. In this paper, a novel compressed sensing method for PACT using an untrained neural network is proposed, which decreases half number of the measured channels and recoveries enough details. This method uses a neural network to reconstruct without the requirement for any additional learning based on the deep image prior. The model can reconstruct the image only using a few detections with gradient descent. Our method can cooperate with other existing regularization, and further improve the quality. In addition, we introduce a shape prior to easily converge the model to the image. We verify the feasibility of untrained network based compressed sensing in PA image reconstruction, and compare this method with a conventional method using total variation minimization. The experimental results show that our proposed method outperforms 32.72% (SSIM) with the traditional compressed sensing method in the same regularization. It could dramatically reduce the requirement for the number of transducers, by sparsely sampling the raw PA data, and improve the quality of PA image significantly.
AS-Net: Fast Photoacoustic Reconstruction with Multi-feature Fusion from Sparse Data
Jan 22, 2021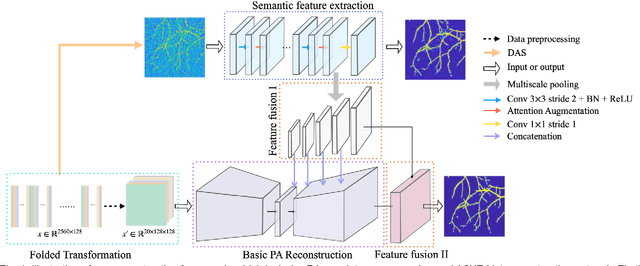
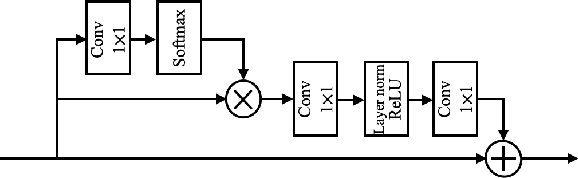
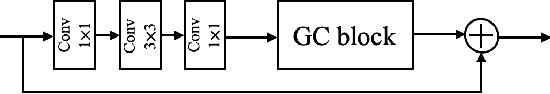
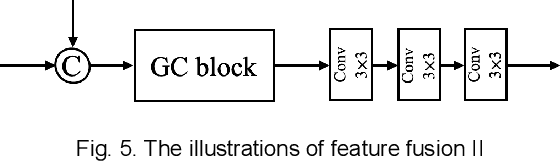
Abstract:Photoacoustic (PA) imaging is a biomedical imaging modality capable of acquiring high contrast images of optical absorption at depths much greater than traditional optical imaging techniques. However, practical instrumentation and geometry limit the number of available acoustic sensors surrounding the imaging target, which results in sparsity of sensor data. Conventional PA image reconstruction methods give severe artifacts when they are applied directly to these sparse data. In this paper, we first employ a novel signal processing method to make sparse PA raw data more suitable for the neural network, and concurrently speeding up image reconstruction. Then we propose Attention Steered Network (AS-Net) for PA reconstruction with multi-feature fusion. AS-Net is validated on different datasets, including simulated photoacoustic data from fundus vasculature phantoms and real data from in vivo fish and mice imaging experiments. Notably, the method is also able to eliminate some artifacts present in the ground-truth for in vivo data. Results demonstrated that our method provides superior reconstructions at a faster speed.
Better Than Ground-truth? Beyond Supervised Learning for Photoacoustic Imaging Reconstruction
Dec 21, 2020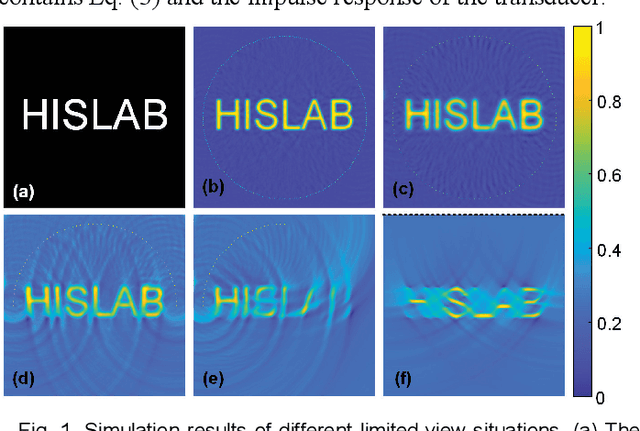
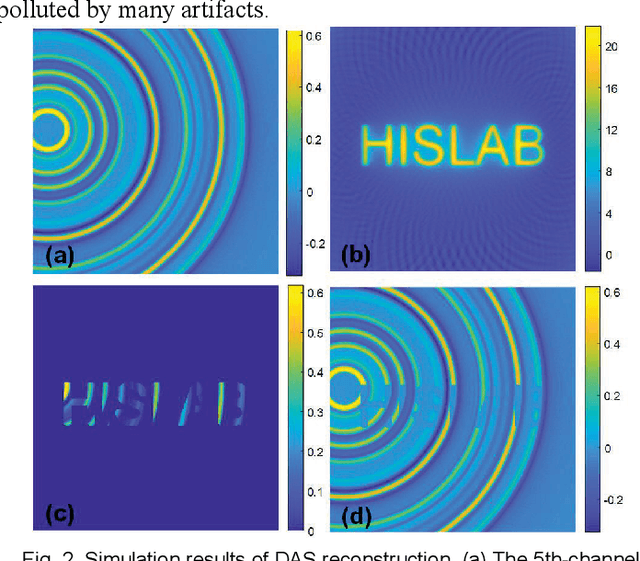
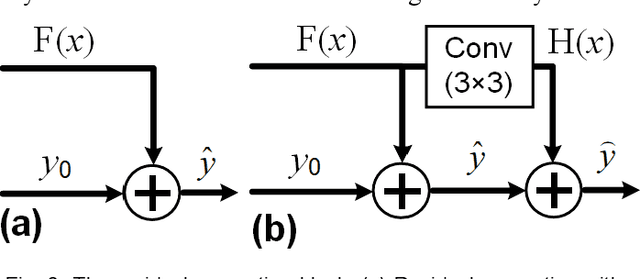
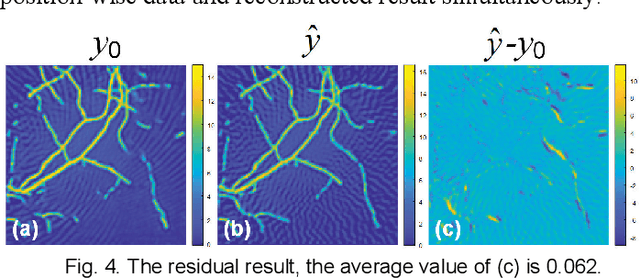
Abstract:Photoacoustic computed tomography (PACT) reconstructs the initial pressure distribution from raw PA signals. Standard reconstruction always induces artifacts using limited-view signals, which are influenced by limited angle coverage of transducers, finite bandwidth, and uncertain heterogeneous biological tissue. Recently, supervised deep learning has been used to overcome limited-view problem that requires ground-truth. However, even full-view sampling still induces artifacts that cannot be used to train the model. It causes a dilemma that we could not acquire perfect ground-truth in practice. To reduce the dependence on the quality of ground-truth, in this paper, for the first time, we propose a beyond supervised reconstruction framework (BSR-Net) based on deep learning to compensate the limited-view issue by feeding limited-view position-wise data. A quarter position-wise data is fed into model and outputs a group full-view data. Specifically, our method introduces a residual structure, which generates beyond supervised reconstruction result, whose artifacts are drastically reduced in the output compared to ground-truth. Moreover, two novel losses are designed to restrain the artifacts. The numerical and in-vivo results have demonstrated the performance of our method to reconstruct the full-view image without artifacts.
Deep Learning Enables Robust and Precise Light Focusing on Treatment Needs
Aug 16, 2020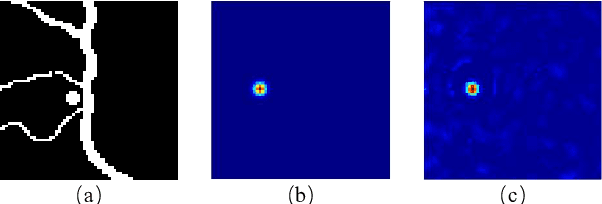

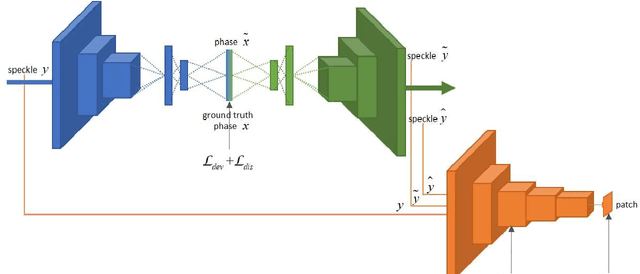
Abstract:If light passes through the body tissues, focusing only on areas where treatment needs, such as tumors, will revolutionize many biomedical imaging and therapy technologies. So how to focus light through deep inhomogeneous tissues overcoming scattering is Holy Grail in biomedical areas. In this paper, we use deep learning to learn and accelerate the process of phase pre-compensation using wavefront shaping. We present an approach (LoftGAN, light only focuses on treatment needs) for learning the relationship between phase domain X and speckle domain Y . Our goal is not just to learn an inverse mapping F:Y->X such that we can know the corresponding X needed for imaging Y like most work, but also to make focusing that is susceptible to disturbances more robust and precise by ensuring that the phase obtained can be forward mapped back to speckle. So we introduce different constraints to enforce F(Y)=X and H(F(Y))=Y with the transmission mapping H:X->Y. Both simulation and physical experiments are performed to investigate the effects of light focusing to demonstrate the effectiveness of our method and comparative experiments prove the crucial improvement of robustness and precision. Codes are available at https://github.com/ChangchunYang/LoftGAN.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge