Gongning Luo
Faculty of Computing, Harbin Institute of Technology, Harbin, China
Fully Kolmogorov-Arnold Deep Model in Medical Image Segmentation
Feb 03, 2026Abstract:Deeply stacked KANs are practically impossible due to high training difficulties and substantial memory requirements. Consequently, existing studies can only incorporate few KAN layers, hindering the comprehensive exploration of KANs. This study overcomes these limitations and introduces the first fully KA-based deep model, demonstrating that KA-based layers can entirely replace traditional architectures in deep learning and achieve superior learning capacity. Specifically, (1) the proposed Share-activation KAN (SaKAN) reformulates Sprecher's variant of Kolmogorov-Arnold representation theorem, which achieves better optimization due to its simplified parameterization and denser training samples, to ease training difficulty, (2) this paper indicates that spline gradients contribute negligibly to training while consuming huge GPU memory, thus proposes the Grad-Free Spline to significantly reduce memory usage and computational overhead. (3) Building on these two innovations, our ALL U-KAN is the first representative implementation of fully KA-based deep model, where the proposed KA and KAonv layers completely replace FC and Conv layers. Extensive evaluations on three medical image segmentation tasks confirm the superiority of the full KA-based architecture compared to partial KA-based and traditional architectures, achieving all higher segmentation accuracy. Compared to directly deeply stacked KAN, ALL U-KAN achieves 10 times reduction in parameter count and reduces memory consumption by more than 20 times, unlocking the new explorations into deep KAN architectures.
FUGC: Benchmarking Semi-Supervised Learning Methods for Cervical Segmentation
Jan 22, 2026Abstract:Accurate segmentation of cervical structures in transvaginal ultrasound (TVS) is critical for assessing the risk of spontaneous preterm birth (PTB), yet the scarcity of labeled data limits the performance of supervised learning approaches. This paper introduces the Fetal Ultrasound Grand Challenge (FUGC), the first benchmark for semi-supervised learning in cervical segmentation, hosted at ISBI 2025. FUGC provides a dataset of 890 TVS images, including 500 training images, 90 validation images, and 300 test images. Methods were evaluated using the Dice Similarity Coefficient (DSC), Hausdorff Distance (HD), and runtime (RT), with a weighted combination of 0.4/0.4/0.2. The challenge attracted 10 teams with 82 participants submitting innovative solutions. The best-performing methods for each individual metric achieved 90.26\% mDSC, 38.88 mHD, and 32.85 ms RT, respectively. FUGC establishes a standardized benchmark for cervical segmentation, demonstrates the efficacy of semi-supervised methods with limited labeled data, and provides a foundation for AI-assisted clinical PTB risk assessment.
Revisiting Data Scaling Law for Medical Segmentation
Nov 17, 2025Abstract:The population loss of trained deep neural networks often exhibits power law scaling with the size of the training dataset, guiding significant performance advancements in deep learning applications. In this study, we focus on the scaling relationship with data size in the context of medical anatomical segmentation, a domain that remains underexplored. We analyze scaling laws for anatomical segmentation across 15 semantic tasks and 4 imaging modalities, demonstrating that larger datasets significantly improve segmentation performance, following similar scaling trends. Motivated by the topological isomorphism in images sharing anatomical structures, we evaluate the impact of deformation-guided augmentation strategies on data scaling laws, specifically random elastic deformation and registration-guided deformation. We also propose a novel, scalable image augmentation approach that generates diffeomorphic mappings from geodesic subspace based on image registration to introduce realistic deformation. Our experimental results demonstrate that both registered and generated deformation-based augmentation considerably enhance data utilization efficiency. The proposed generated deformation method notably achieves superior performance and accelerated convergence, surpassing standard power law scaling trends without requiring additional data. Overall, this work provides insights into the understanding of segmentation scalability and topological variation impact in medical imaging, thereby leading to more efficient model development with reduced annotation and computational costs.
Ambiguity-aware Truncated Flow Matching for Ambiguous Medical Image Segmentation
Nov 10, 2025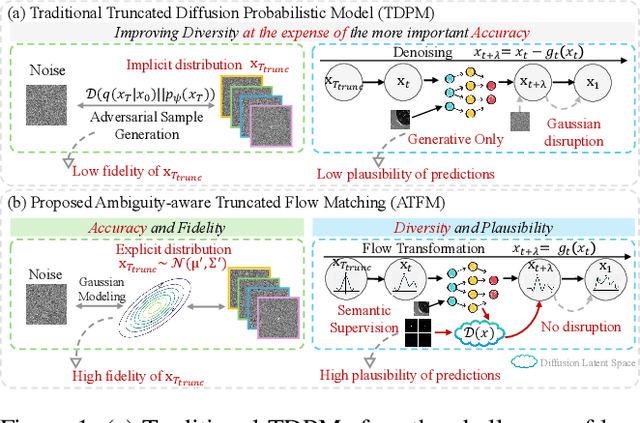
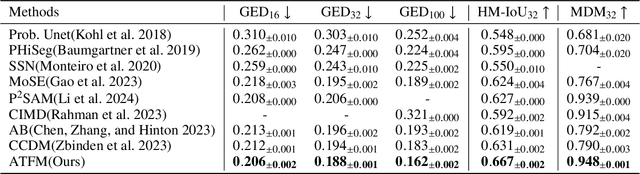
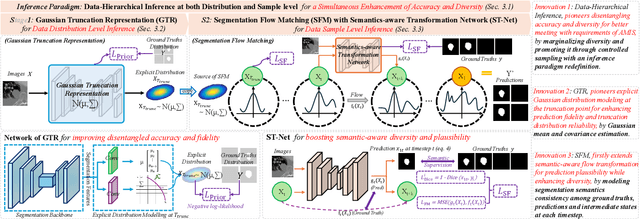
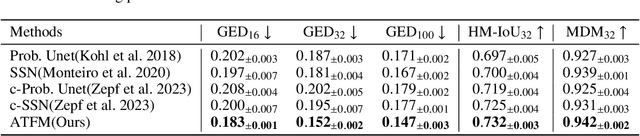
Abstract:A simultaneous enhancement of accuracy and diversity of predictions remains a challenge in ambiguous medical image segmentation (AMIS) due to the inherent trade-offs. While truncated diffusion probabilistic models (TDPMs) hold strong potential with a paradigm optimization, existing TDPMs suffer from entangled accuracy and diversity of predictions with insufficient fidelity and plausibility. To address the aforementioned challenges, we propose Ambiguity-aware Truncated Flow Matching (ATFM), which introduces a novel inference paradigm and dedicated model components. Firstly, we propose Data-Hierarchical Inference, a redefinition of AMIS-specific inference paradigm, which enhances accuracy and diversity at data-distribution and data-sample level, respectively, for an effective disentanglement. Secondly, Gaussian Truncation Representation (GTR) is introduced to enhance both fidelity of predictions and reliability of truncation distribution, by explicitly modeling it as a Gaussian distribution at $T_{\text{trunc}}$ instead of using sampling-based approximations.Thirdly, Segmentation Flow Matching (SFM) is proposed to enhance the plausibility of diverse predictions by extending semantic-aware flow transformation in Flow Matching (FM). Comprehensive evaluations on LIDC and ISIC3 datasets demonstrate that ATFM outperforms SOTA methods and simultaneously achieves a more efficient inference. ATFM improves GED and HM-IoU by up to $12\%$ and $7.3\%$ compared to advanced methods.
Structure and Smoothness Constrained Dual Networks for MR Bias Field Correction
Jul 02, 2025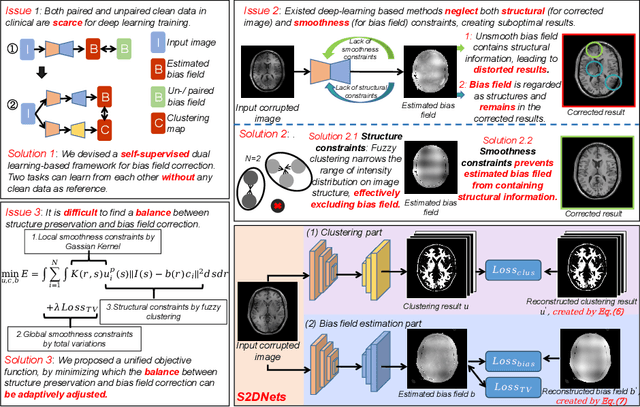
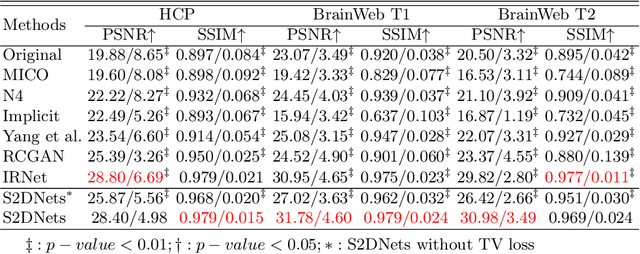
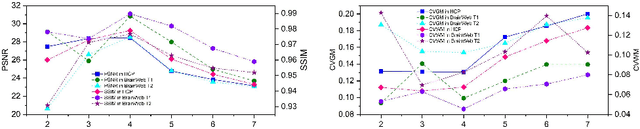
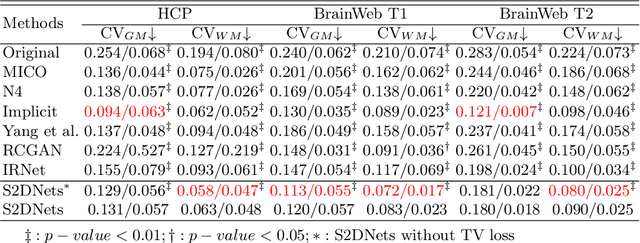
Abstract:MR imaging techniques are of great benefit to disease diagnosis. However, due to the limitation of MR devices, significant intensity inhomogeneity often exists in imaging results, which impedes both qualitative and quantitative medical analysis. Recently, several unsupervised deep learning-based models have been proposed for MR image improvement. However, these models merely concentrate on global appearance learning, and neglect constraints from image structures and smoothness of bias field, leading to distorted corrected results. In this paper, novel structure and smoothness constrained dual networks, named S2DNets, are proposed aiming to self-supervised bias field correction. S2DNets introduce piece-wise structural constraints and smoothness of bias field for network training to effectively remove non-uniform intensity and retain much more structural details. Extensive experiments executed on both clinical and simulated MR datasets show that the proposed model outperforms other conventional and deep learning-based models. In addition to comparison on visual metrics, downstream MR image segmentation tasks are also used to evaluate the impact of the proposed model. The source code is available at: https://github.com/LeongDong/S2DNets}{https://github.com/LeongDong/S2DNets.
* 11 pages, 3 figures, accepted by MICCAI
Finding Local Diffusion Schrödinger Bridge using Kolmogorov-Arnold Network
Feb 27, 2025Abstract:In image generation, Schr\"odinger Bridge (SB)-based methods theoretically enhance the efficiency and quality compared to the diffusion models by finding the least costly path between two distributions. However, they are computationally expensive and time-consuming when applied to complex image data. The reason is that they focus on fitting globally optimal paths in high-dimensional spaces, directly generating images as next step on the path using complex networks through self-supervised training, which typically results in a gap with the global optimum. Meanwhile, most diffusion models are in the same path subspace generated by weights $f_A(t)$ and $f_B(t)$, as they follow the paradigm ($x_t = f_A(t)x_{Img} + f_B(t)\epsilon$). To address the limitations of SB-based methods, this paper proposes for the first time to find local Diffusion Schr\"odinger Bridges (LDSB) in the diffusion path subspace, which strengthens the connection between the SB problem and diffusion models. Specifically, our method optimizes the diffusion paths using Kolmogorov-Arnold Network (KAN), which has the advantage of resistance to forgetting and continuous output. The experiment shows that our LDSB significantly improves the quality and efficiency of image generation using the same pre-trained denoising network and the KAN for optimising is only less than 0.1MB. The FID metric is reduced by \textbf{more than 15\%}, especially with a reduction of 48.50\% when NFE of DDIM is $5$ for the CelebA dataset. Code is available at https://github.com/Qiu-XY/LDSB.
Tumor Detection, Segmentation and Classification Challenge on Automated 3D Breast Ultrasound: The TDSC-ABUS Challenge
Jan 26, 2025



Abstract:Breast cancer is one of the most common causes of death among women worldwide. Early detection helps in reducing the number of deaths. Automated 3D Breast Ultrasound (ABUS) is a newer approach for breast screening, which has many advantages over handheld mammography such as safety, speed, and higher detection rate of breast cancer. Tumor detection, segmentation, and classification are key components in the analysis of medical images, especially challenging in the context of 3D ABUS due to the significant variability in tumor size and shape, unclear tumor boundaries, and a low signal-to-noise ratio. The lack of publicly accessible, well-labeled ABUS datasets further hinders the advancement of systems for breast tumor analysis. Addressing this gap, we have organized the inaugural Tumor Detection, Segmentation, and Classification Challenge on Automated 3D Breast Ultrasound 2023 (TDSC-ABUS2023). This initiative aims to spearhead research in this field and create a definitive benchmark for tasks associated with 3D ABUS image analysis. In this paper, we summarize the top-performing algorithms from the challenge and provide critical analysis for ABUS image examination. We offer the TDSC-ABUS challenge as an open-access platform at https://tdsc-abus2023.grand-challenge.org/ to benchmark and inspire future developments in algorithmic research.
MedFILIP: Medical Fine-grained Language-Image Pre-training
Jan 18, 2025Abstract:Medical vision-language pretraining (VLP) that leverages naturally-paired medical image-report data is crucial for medical image analysis. However, existing methods struggle to accurately characterize associations between images and diseases, leading to inaccurate or incomplete diagnostic results. In this work, we propose MedFILIP, a fine-grained VLP model, introduces medical image-specific knowledge through contrastive learning, specifically: 1) An information extractor based on a large language model is proposed to decouple comprehensive disease details from reports, which excels in extracting disease deals through flexible prompt engineering, thereby effectively reducing text complexity while retaining rich information at a tiny cost. 2) A knowledge injector is proposed to construct relationships between categories and visual attributes, which help the model to make judgments based on image features, and fosters knowledge extrapolation to unfamiliar disease categories. 3) A semantic similarity matrix based on fine-grained annotations is proposed, providing smoother, information-richer labels, thus allowing fine-grained image-text alignment. 4) We validate MedFILIP on numerous datasets, e.g., RSNA-Pneumonia, NIH ChestX-ray14, VinBigData, and COVID-19. For single-label, multi-label, and fine-grained classification, our model achieves state-of-the-art performance, the classification accuracy has increased by a maximum of 6.69\%. The code is available in https://github.com/PerceptionComputingLab/MedFILIP.
Efficient MedSAMs: Segment Anything in Medical Images on Laptop
Dec 20, 2024


Abstract:Promptable segmentation foundation models have emerged as a transformative approach to addressing the diverse needs in medical images, but most existing models require expensive computing, posing a big barrier to their adoption in clinical practice. In this work, we organized the first international competition dedicated to promptable medical image segmentation, featuring a large-scale dataset spanning nine common imaging modalities from over 20 different institutions. The top teams developed lightweight segmentation foundation models and implemented an efficient inference pipeline that substantially reduced computational requirements while maintaining state-of-the-art segmentation accuracy. Moreover, the post-challenge phase advanced the algorithms through the design of performance booster and reproducibility tasks, resulting in improved algorithms and validated reproducibility of the winning solution. Furthermore, the best-performing algorithms have been incorporated into the open-source software with a user-friendly interface to facilitate clinical adoption. The data and code are publicly available to foster the further development of medical image segmentation foundation models and pave the way for impactful real-world applications.
Improving Representation of High-frequency Components for Medical Foundation Models
Jul 26, 2024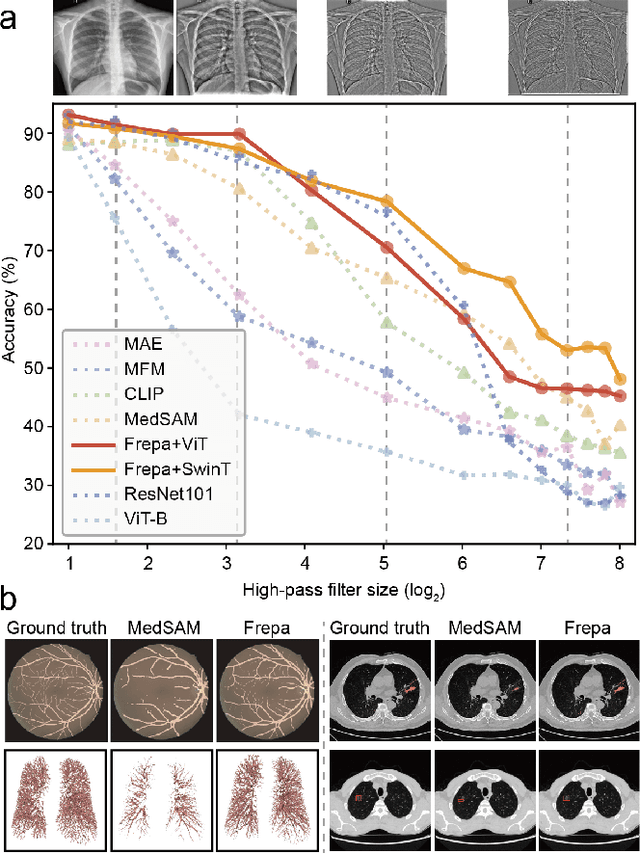

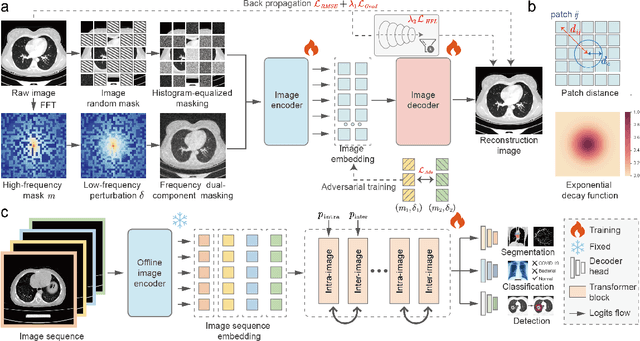

Abstract:Foundation models have recently attracted significant attention for their impressive generalizability across diverse downstream tasks. However, these models are demonstrated to exhibit great limitations in representing high-frequency components and fine-grained details. In many medical imaging tasks, the precise representation of such information is crucial due to the inherently intricate anatomical structures, sub-visual features, and complex boundaries involved. Consequently, the limited representation of prevalent foundation models can result in significant performance degradation or even failure in these tasks. To address these challenges, we propose a novel pretraining strategy, named Frequency-advanced Representation Autoencoder (Frepa). Through high-frequency masking and low-frequency perturbation combined with adversarial learning, Frepa encourages the encoder to effectively represent and preserve high-frequency components in the image embeddings. Additionally, we introduce an innovative histogram-equalized image masking strategy, extending the Masked Autoencoder approach beyond ViT to other architectures such as Swin Transformer and convolutional networks. We develop Frepa across nine medical modalities and validate it on 32 downstream tasks for both 2D images and 3D volume data. Without fine-tuning, Frepa can outperform other self-supervised pretraining methods and, in some cases, even surpasses task-specific trained models. This improvement is particularly significant for tasks involving fine-grained details, such as achieving up to a +15% increase in DSC for retina vessel segmentation and a +7% increase in IoU for lung nodule detection. Further experiments quantitatively reveal that Frepa enables superior high-frequency representations and preservation in the embeddings, underscoring its potential for developing more generalized and universal medical image foundation models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge