Suyu Dong
College of Computer and Control Engineering, Northeast Forestry University, Harbin, China
Ambiguity-aware Truncated Flow Matching for Ambiguous Medical Image Segmentation
Nov 10, 2025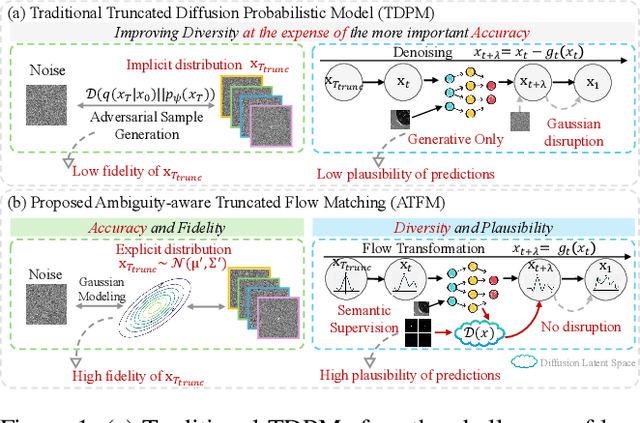
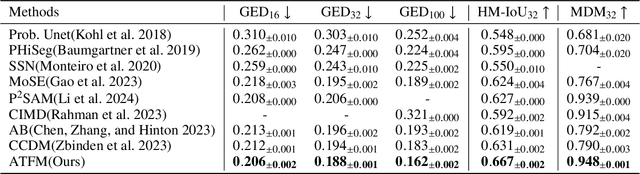
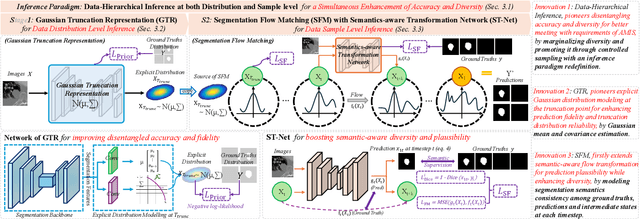
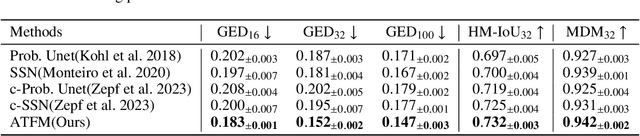
Abstract:A simultaneous enhancement of accuracy and diversity of predictions remains a challenge in ambiguous medical image segmentation (AMIS) due to the inherent trade-offs. While truncated diffusion probabilistic models (TDPMs) hold strong potential with a paradigm optimization, existing TDPMs suffer from entangled accuracy and diversity of predictions with insufficient fidelity and plausibility. To address the aforementioned challenges, we propose Ambiguity-aware Truncated Flow Matching (ATFM), which introduces a novel inference paradigm and dedicated model components. Firstly, we propose Data-Hierarchical Inference, a redefinition of AMIS-specific inference paradigm, which enhances accuracy and diversity at data-distribution and data-sample level, respectively, for an effective disentanglement. Secondly, Gaussian Truncation Representation (GTR) is introduced to enhance both fidelity of predictions and reliability of truncation distribution, by explicitly modeling it as a Gaussian distribution at $T_{\text{trunc}}$ instead of using sampling-based approximations.Thirdly, Segmentation Flow Matching (SFM) is proposed to enhance the plausibility of diverse predictions by extending semantic-aware flow transformation in Flow Matching (FM). Comprehensive evaluations on LIDC and ISIC3 datasets demonstrate that ATFM outperforms SOTA methods and simultaneously achieves a more efficient inference. ATFM improves GED and HM-IoU by up to $12\%$ and $7.3\%$ compared to advanced methods.
Structure and Smoothness Constrained Dual Networks for MR Bias Field Correction
Jul 02, 2025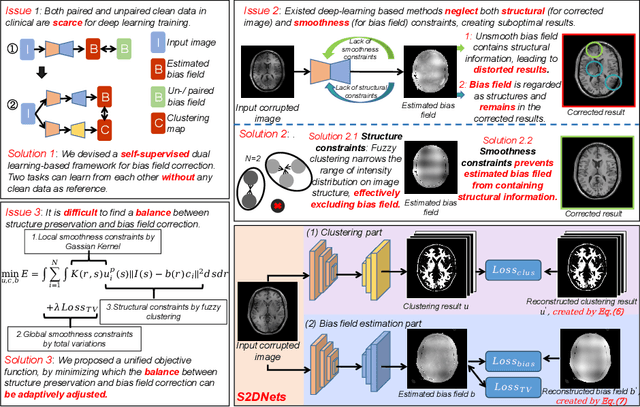
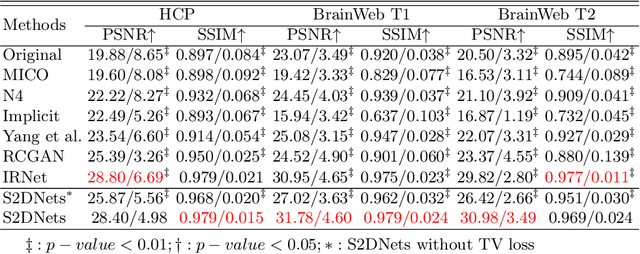
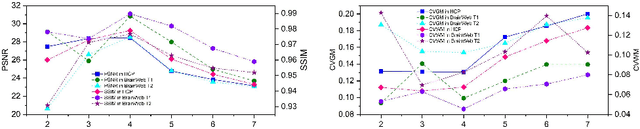
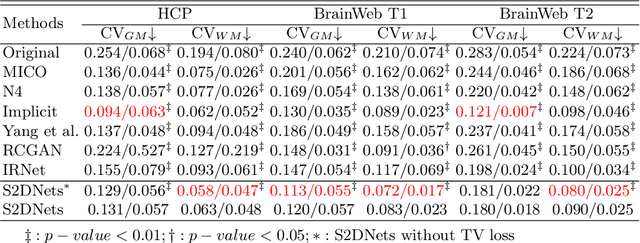
Abstract:MR imaging techniques are of great benefit to disease diagnosis. However, due to the limitation of MR devices, significant intensity inhomogeneity often exists in imaging results, which impedes both qualitative and quantitative medical analysis. Recently, several unsupervised deep learning-based models have been proposed for MR image improvement. However, these models merely concentrate on global appearance learning, and neglect constraints from image structures and smoothness of bias field, leading to distorted corrected results. In this paper, novel structure and smoothness constrained dual networks, named S2DNets, are proposed aiming to self-supervised bias field correction. S2DNets introduce piece-wise structural constraints and smoothness of bias field for network training to effectively remove non-uniform intensity and retain much more structural details. Extensive experiments executed on both clinical and simulated MR datasets show that the proposed model outperforms other conventional and deep learning-based models. In addition to comparison on visual metrics, downstream MR image segmentation tasks are also used to evaluate the impact of the proposed model. The source code is available at: https://github.com/LeongDong/S2DNets}{https://github.com/LeongDong/S2DNets.
* 11 pages, 3 figures, accepted by MICCAI
Tumor Detection, Segmentation and Classification Challenge on Automated 3D Breast Ultrasound: The TDSC-ABUS Challenge
Jan 26, 2025



Abstract:Breast cancer is one of the most common causes of death among women worldwide. Early detection helps in reducing the number of deaths. Automated 3D Breast Ultrasound (ABUS) is a newer approach for breast screening, which has many advantages over handheld mammography such as safety, speed, and higher detection rate of breast cancer. Tumor detection, segmentation, and classification are key components in the analysis of medical images, especially challenging in the context of 3D ABUS due to the significant variability in tumor size and shape, unclear tumor boundaries, and a low signal-to-noise ratio. The lack of publicly accessible, well-labeled ABUS datasets further hinders the advancement of systems for breast tumor analysis. Addressing this gap, we have organized the inaugural Tumor Detection, Segmentation, and Classification Challenge on Automated 3D Breast Ultrasound 2023 (TDSC-ABUS2023). This initiative aims to spearhead research in this field and create a definitive benchmark for tasks associated with 3D ABUS image analysis. In this paper, we summarize the top-performing algorithms from the challenge and provide critical analysis for ABUS image examination. We offer the TDSC-ABUS challenge as an open-access platform at https://tdsc-abus2023.grand-challenge.org/ to benchmark and inspire future developments in algorithmic research.
MedFILIP: Medical Fine-grained Language-Image Pre-training
Jan 18, 2025Abstract:Medical vision-language pretraining (VLP) that leverages naturally-paired medical image-report data is crucial for medical image analysis. However, existing methods struggle to accurately characterize associations between images and diseases, leading to inaccurate or incomplete diagnostic results. In this work, we propose MedFILIP, a fine-grained VLP model, introduces medical image-specific knowledge through contrastive learning, specifically: 1) An information extractor based on a large language model is proposed to decouple comprehensive disease details from reports, which excels in extracting disease deals through flexible prompt engineering, thereby effectively reducing text complexity while retaining rich information at a tiny cost. 2) A knowledge injector is proposed to construct relationships between categories and visual attributes, which help the model to make judgments based on image features, and fosters knowledge extrapolation to unfamiliar disease categories. 3) A semantic similarity matrix based on fine-grained annotations is proposed, providing smoother, information-richer labels, thus allowing fine-grained image-text alignment. 4) We validate MedFILIP on numerous datasets, e.g., RSNA-Pneumonia, NIH ChestX-ray14, VinBigData, and COVID-19. For single-label, multi-label, and fine-grained classification, our model achieves state-of-the-art performance, the classification accuracy has increased by a maximum of 6.69\%. The code is available in https://github.com/PerceptionComputingLab/MedFILIP.
Efficient automatic segmentation for multi-level pulmonary arteries: The PARSE challenge
Apr 07, 2023



Abstract:Efficient automatic segmentation of multi-level (i.e. main and branch) pulmonary arteries (PA) in CTPA images plays a significant role in clinical applications. However, most existing methods concentrate only on main PA or branch PA segmentation separately and ignore segmentation efficiency. Besides, there is no public large-scale dataset focused on PA segmentation, which makes it highly challenging to compare the different methods. To benchmark multi-level PA segmentation algorithms, we organized the first \textbf{P}ulmonary \textbf{AR}tery \textbf{SE}gmentation (PARSE) challenge. On the one hand, we focus on both the main PA and the branch PA segmentation. On the other hand, for better clinical application, we assign the same score weight to segmentation efficiency (mainly running time and GPU memory consumption during inference) while ensuring PA segmentation accuracy. We present a summary of the top algorithms and offer some suggestions for efficient and accurate multi-level PA automatic segmentation. We provide the PARSE challenge as open-access for the community to benchmark future algorithm developments at \url{https://parse2022.grand-challenge.org/Parse2022/}.
VoxelAtlasGAN: 3D Left Ventricle Segmentation on Echocardiography with Atlas Guided Generation and Voxel-to-voxel Discrimination
Jun 10, 2018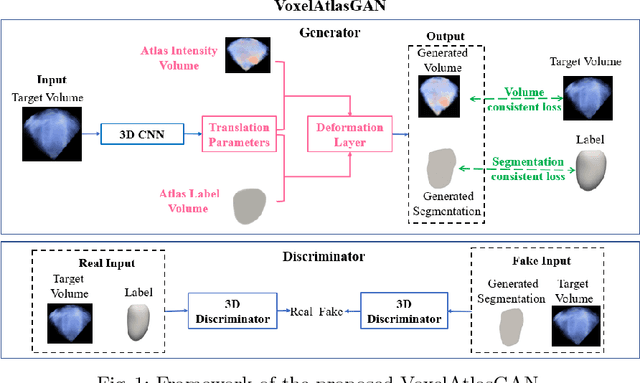

Abstract:3D left ventricle (LV) segmentation on echocardiography is very important for diagnosis and treatment of cardiac disease. It is not only because of that echocardiography is a real-time imaging technology and widespread in clinical application, but also because of that LV segmentation on 3D echocardiography can provide more full volume information of heart than LV segmentation on 2D echocardiography. However, 3D LV segmentation on echocardiography is still an open and challenging task owing to the lower contrast, higher noise and data dimensionality, limited annotation of 3D echocardiography. In this paper, we proposed a novel real-time framework, i.e., VoxelAtlasGAN, for 3D LV segmentation on 3D echocardiography. This framework has three contributions: 1) It is based on voxel-to-voxel conditional generative adversarial nets (cGAN). For the first time, cGAN is used for 3D LV segmentation on echocardiography. And cGAN advantageously fuses substantial 3D spatial context information from 3D echocardiography by self-learning structured loss; 2) For the first time, it embeds the atlas into an end-to-end optimization framework, which uses 3D LV atlas as a powerful prior knowledge to improve the inference speed, address the lower contrast and the limited annotation problems of 3D echocardiography; 3) It combines traditional discrimination loss and the new proposed consistent constraint, which further improves the generalization of the proposed framework. VoxelAtlasGAN was validated on 60 subjects on 3D echocardiography and it achieved satisfactory segmentation results and high inference speed. The mean surface distance is 1.85 mm, the mean hausdorff surface distance is 7.26 mm, mean dice is 0.953, the correlation of EF is 0.918, and the mean inference speed is 0.1s. These results have demonstrated that our proposed method has great potential for clinical application
Multi-views Fusion CNN for Left Ventricular Volumes Estimation on Cardiac MR Images
Apr 09, 2018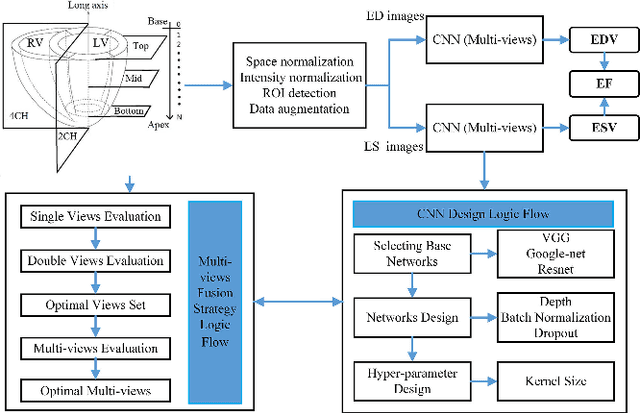
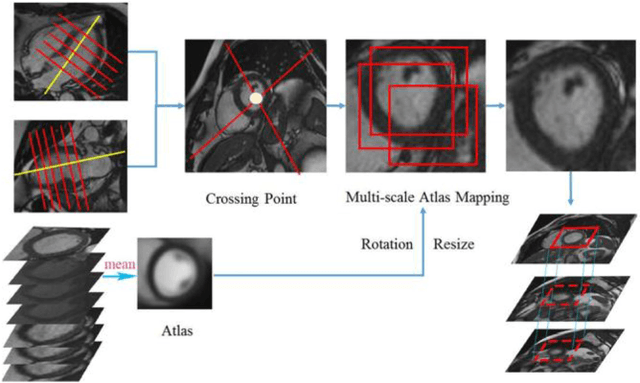
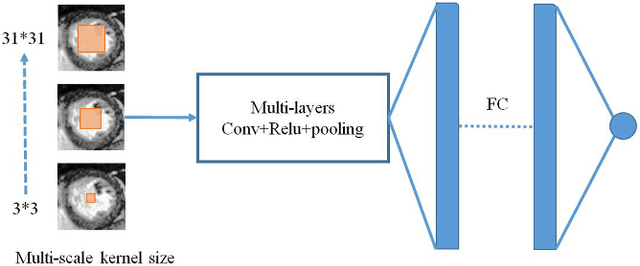
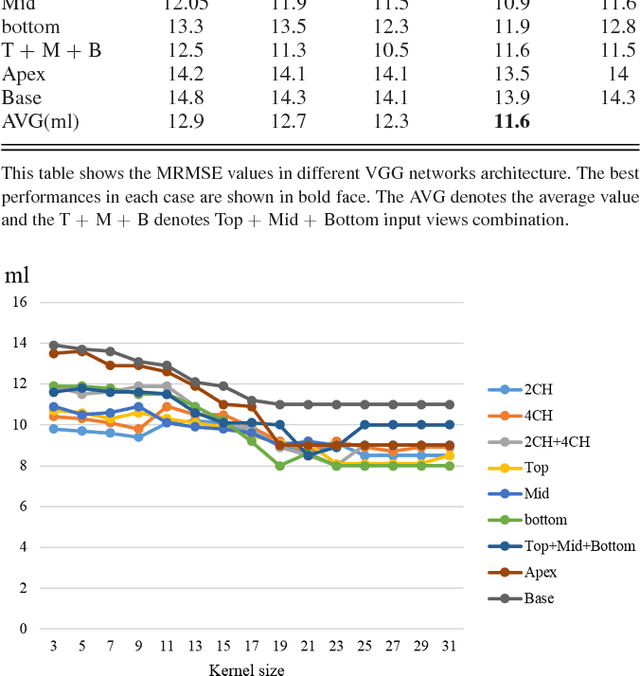
Abstract:Left ventricular (LV) volumes estimation is a critical procedure for cardiac disease diagnosis. The objective of this paper is to address direct LV volumes prediction task. Methods: In this paper, we propose a direct volumes prediction method based on the end-to-end deep convolutional neural networks (CNN). We study the end-to-end LV volumes prediction method in items of the data preprocessing, networks structure, and multi-views fusion strategy. The main contributions of this paper are the following aspects. First, we propose a new data preprocessing method on cardiac magnetic resonance (CMR). Second, we propose a new networks structure for end-to-end LV volumes estimation. Third, we explore the representational capacity of different slices, and propose a fusion strategy to improve the prediction accuracy. Results: The evaluation results show that the proposed method outperforms other state-of-the-art LV volumes estimation methods on the open accessible benchmark datasets. The clinical indexes derived from the predicted volumes agree well with the ground truth (EDV: R2=0.974, RMSE=9.6ml; ESV: R2=0.976, RMSE=7.1ml; EF: R2=0.828, RMSE =4.71%). Conclusion: Experimental results prove that the proposed method may be useful for LV volumes prediction task. Significance: The proposed method not only has application potential for cardiac diseases screening for large-scale CMR data, but also can be extended to other medical image research fields
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge