Fanding Li
Faculty of Computing, Harbin Institute of Technology, Harbin, China
Ambiguity-aware Truncated Flow Matching for Ambiguous Medical Image Segmentation
Nov 10, 2025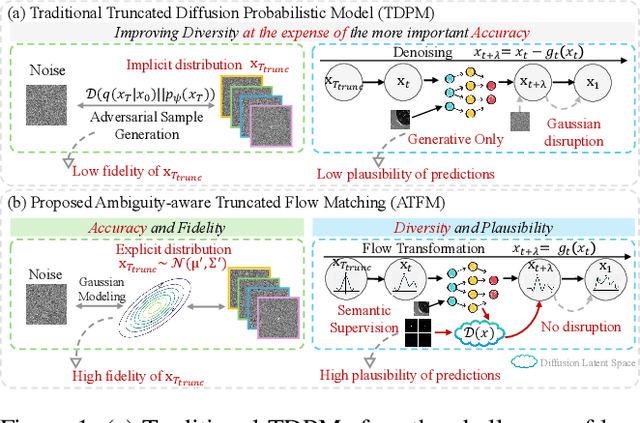
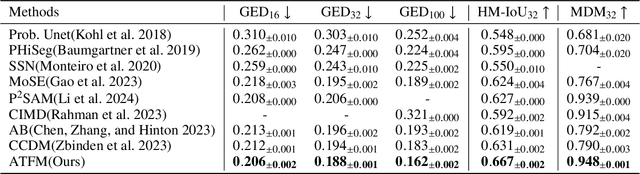
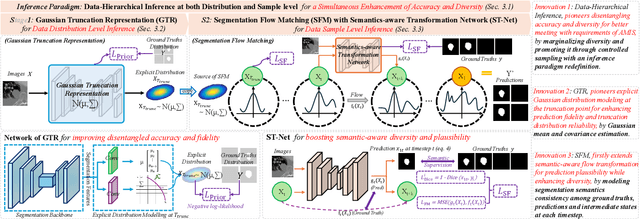
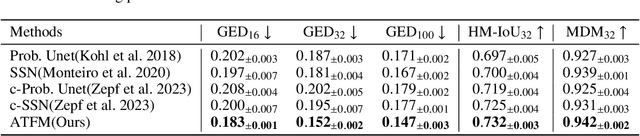
Abstract:A simultaneous enhancement of accuracy and diversity of predictions remains a challenge in ambiguous medical image segmentation (AMIS) due to the inherent trade-offs. While truncated diffusion probabilistic models (TDPMs) hold strong potential with a paradigm optimization, existing TDPMs suffer from entangled accuracy and diversity of predictions with insufficient fidelity and plausibility. To address the aforementioned challenges, we propose Ambiguity-aware Truncated Flow Matching (ATFM), which introduces a novel inference paradigm and dedicated model components. Firstly, we propose Data-Hierarchical Inference, a redefinition of AMIS-specific inference paradigm, which enhances accuracy and diversity at data-distribution and data-sample level, respectively, for an effective disentanglement. Secondly, Gaussian Truncation Representation (GTR) is introduced to enhance both fidelity of predictions and reliability of truncation distribution, by explicitly modeling it as a Gaussian distribution at $T_{\text{trunc}}$ instead of using sampling-based approximations.Thirdly, Segmentation Flow Matching (SFM) is proposed to enhance the plausibility of diverse predictions by extending semantic-aware flow transformation in Flow Matching (FM). Comprehensive evaluations on LIDC and ISIC3 datasets demonstrate that ATFM outperforms SOTA methods and simultaneously achieves a more efficient inference. ATFM improves GED and HM-IoU by up to $12\%$ and $7.3\%$ compared to advanced methods.
Finding Local Diffusion Schrödinger Bridge using Kolmogorov-Arnold Network
Feb 27, 2025Abstract:In image generation, Schr\"odinger Bridge (SB)-based methods theoretically enhance the efficiency and quality compared to the diffusion models by finding the least costly path between two distributions. However, they are computationally expensive and time-consuming when applied to complex image data. The reason is that they focus on fitting globally optimal paths in high-dimensional spaces, directly generating images as next step on the path using complex networks through self-supervised training, which typically results in a gap with the global optimum. Meanwhile, most diffusion models are in the same path subspace generated by weights $f_A(t)$ and $f_B(t)$, as they follow the paradigm ($x_t = f_A(t)x_{Img} + f_B(t)\epsilon$). To address the limitations of SB-based methods, this paper proposes for the first time to find local Diffusion Schr\"odinger Bridges (LDSB) in the diffusion path subspace, which strengthens the connection between the SB problem and diffusion models. Specifically, our method optimizes the diffusion paths using Kolmogorov-Arnold Network (KAN), which has the advantage of resistance to forgetting and continuous output. The experiment shows that our LDSB significantly improves the quality and efficiency of image generation using the same pre-trained denoising network and the KAN for optimising is only less than 0.1MB. The FID metric is reduced by \textbf{more than 15\%}, especially with a reduction of 48.50\% when NFE of DDIM is $5$ for the CelebA dataset. Code is available at https://github.com/Qiu-XY/LDSB.
MedFILIP: Medical Fine-grained Language-Image Pre-training
Jan 18, 2025Abstract:Medical vision-language pretraining (VLP) that leverages naturally-paired medical image-report data is crucial for medical image analysis. However, existing methods struggle to accurately characterize associations between images and diseases, leading to inaccurate or incomplete diagnostic results. In this work, we propose MedFILIP, a fine-grained VLP model, introduces medical image-specific knowledge through contrastive learning, specifically: 1) An information extractor based on a large language model is proposed to decouple comprehensive disease details from reports, which excels in extracting disease deals through flexible prompt engineering, thereby effectively reducing text complexity while retaining rich information at a tiny cost. 2) A knowledge injector is proposed to construct relationships between categories and visual attributes, which help the model to make judgments based on image features, and fosters knowledge extrapolation to unfamiliar disease categories. 3) A semantic similarity matrix based on fine-grained annotations is proposed, providing smoother, information-richer labels, thus allowing fine-grained image-text alignment. 4) We validate MedFILIP on numerous datasets, e.g., RSNA-Pneumonia, NIH ChestX-ray14, VinBigData, and COVID-19. For single-label, multi-label, and fine-grained classification, our model achieves state-of-the-art performance, the classification accuracy has increased by a maximum of 6.69\%. The code is available in https://github.com/PerceptionComputingLab/MedFILIP.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge