Henggui Zhang
Weakly Supervised Arrhythmia Detection Based on Deep Convolutional Neural Network
Dec 10, 2020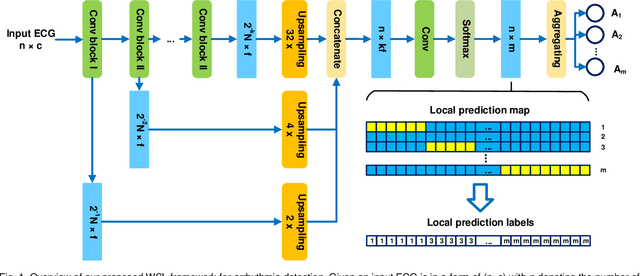
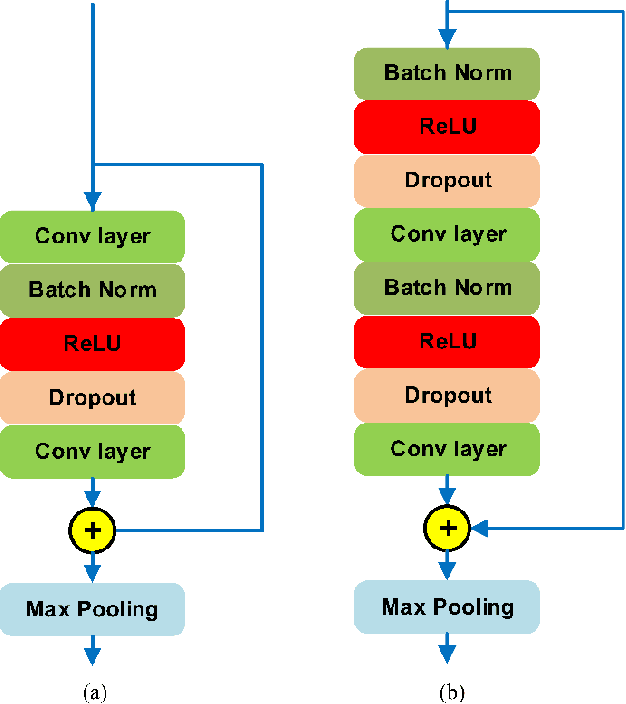
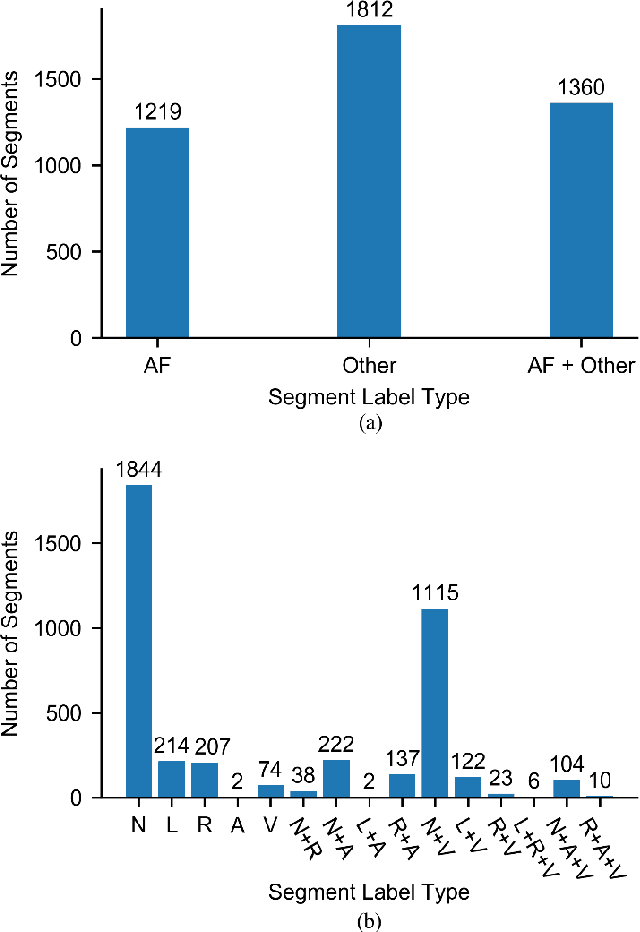
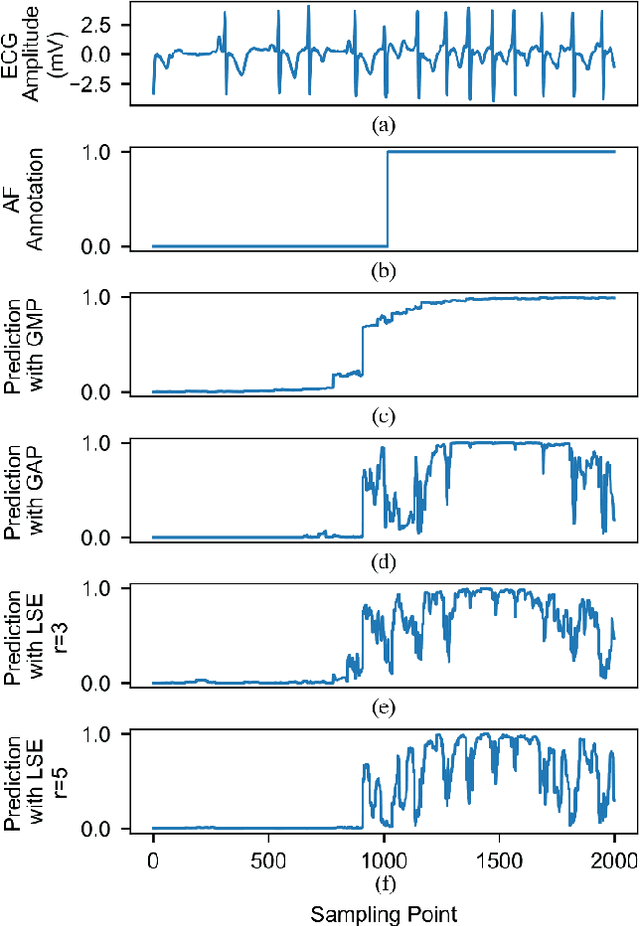
Abstract:Supervised deep learning has been widely used in the studies of automatic ECG classification, which largely benefits from sufficient annotation of large datasets. However, most of the existing large ECG datasets are roughly annotated, so the classification model trained on them can only detect the existence of abnormalities in a whole recording, but cannot determine their exact occurrence time. In addition, it may take huge time and economic cost to construct a fine-annotated ECG dataset. Therefore, this study proposes weakly supervised deep learning models for detecting abnormal ECG events and their occurrence time. The available supervision information for the models is limited to the event types in an ECG record, excluding the specific occurring time of each event. By leverage of feature locality of deep convolution neural network, the models first make predictions based on the local features, and then aggregate the local predictions to infer the existence of each event during the whole record. Through training, the local predictions are expected to reflect the specific occurring time of each event. To test their potentials, we apply the models for detecting cardiac rhythmic and morphological arrhythmias by using the AFDB and MITDB datasets, respectively. The results show that the models achieve beat-level accuracies of 99.09% in detecting atrial fibrillation, and 99.13% in detecting morphological arrhythmias, which are comparable to that of fully supervised learning models, demonstrating their effectiveness. The local prediction maps revealed by this method are also helpful to analyze and diagnose the decision logic of record-level classification models.
Automatic Detection of ECG Abnormalities by using an Ensemble of Deep Residual Networks with Attention
Aug 27, 2019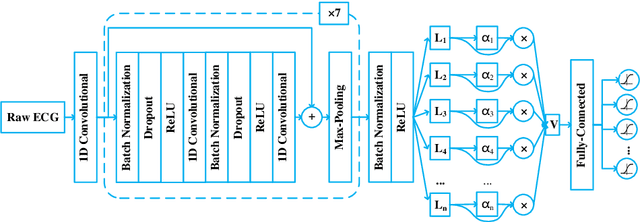
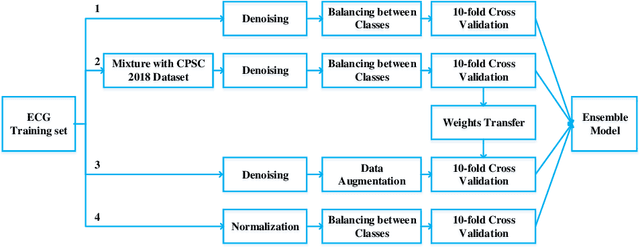
Abstract:Heart disease is one of the most common diseases causing morbidity and mortality. Electrocardiogram (ECG) has been widely used for diagnosing heart diseases for its simplicity and non-invasive property. Automatic ECG analyzing technologies are expected to reduce human working load and increase diagnostic efficacy. However, there are still some challenges to be addressed for achieving this goal. In this study, we develop an algorithm to identify multiple abnormalities from 12-lead ECG recordings. In the algorithm pipeline, several preprocessing methods are firstly applied on the ECG data for denoising, augmentation and balancing recording numbers of variant classes. In consideration of efficiency and consistency of data length, the recordings are padded or truncated into a medium length, where the padding/truncating time windows are selected randomly to sup-press overfitting. Then, the ECGs are used to train deep neural network (DNN) models with a novel structure that combines a deep residual network with an attention mechanism. Finally, an ensemble model is built based on these trained models to make predictions on the test data set. Our method is evaluated based on the test set of the First China ECG Intelligent Competition dataset by using the F1 metric that is regarded as the harmonic mean between the precision and recall. The resultant overall F1 score of the algorithm is 0.875, showing a promising performance and potential for practical use.
VoxelAtlasGAN: 3D Left Ventricle Segmentation on Echocardiography with Atlas Guided Generation and Voxel-to-voxel Discrimination
Jun 10, 2018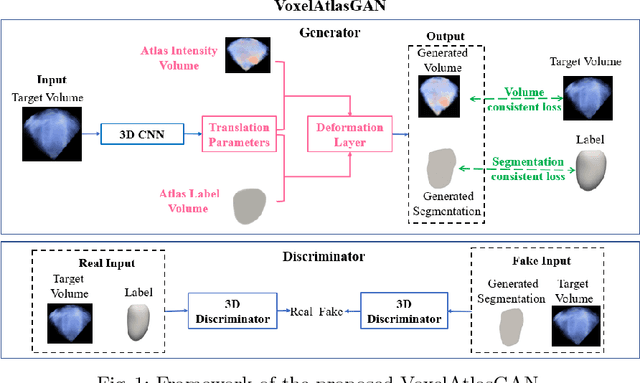

Abstract:3D left ventricle (LV) segmentation on echocardiography is very important for diagnosis and treatment of cardiac disease. It is not only because of that echocardiography is a real-time imaging technology and widespread in clinical application, but also because of that LV segmentation on 3D echocardiography can provide more full volume information of heart than LV segmentation on 2D echocardiography. However, 3D LV segmentation on echocardiography is still an open and challenging task owing to the lower contrast, higher noise and data dimensionality, limited annotation of 3D echocardiography. In this paper, we proposed a novel real-time framework, i.e., VoxelAtlasGAN, for 3D LV segmentation on 3D echocardiography. This framework has three contributions: 1) It is based on voxel-to-voxel conditional generative adversarial nets (cGAN). For the first time, cGAN is used for 3D LV segmentation on echocardiography. And cGAN advantageously fuses substantial 3D spatial context information from 3D echocardiography by self-learning structured loss; 2) For the first time, it embeds the atlas into an end-to-end optimization framework, which uses 3D LV atlas as a powerful prior knowledge to improve the inference speed, address the lower contrast and the limited annotation problems of 3D echocardiography; 3) It combines traditional discrimination loss and the new proposed consistent constraint, which further improves the generalization of the proposed framework. VoxelAtlasGAN was validated on 60 subjects on 3D echocardiography and it achieved satisfactory segmentation results and high inference speed. The mean surface distance is 1.85 mm, the mean hausdorff surface distance is 7.26 mm, mean dice is 0.953, the correlation of EF is 0.918, and the mean inference speed is 0.1s. These results have demonstrated that our proposed method has great potential for clinical application
Multi-views Fusion CNN for Left Ventricular Volumes Estimation on Cardiac MR Images
Apr 09, 2018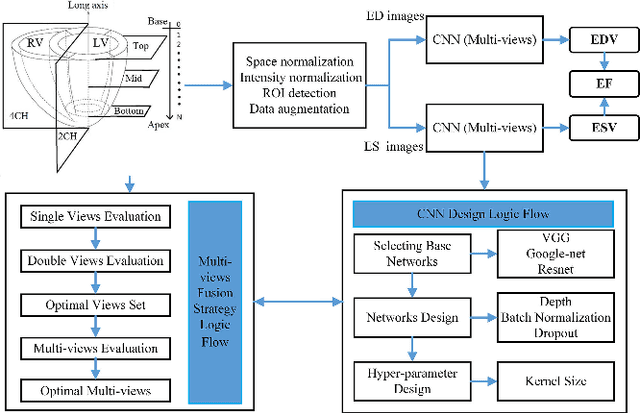
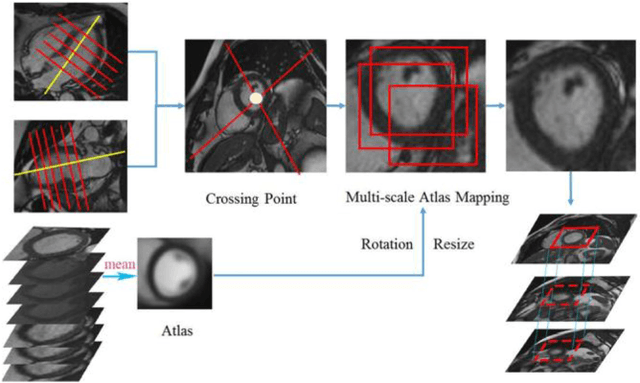
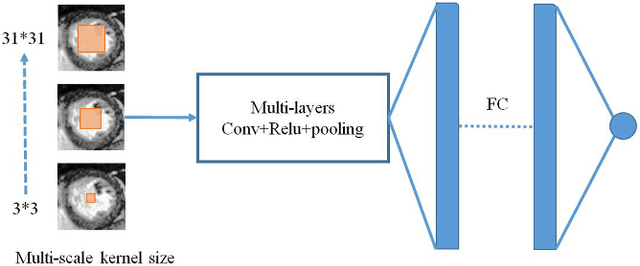
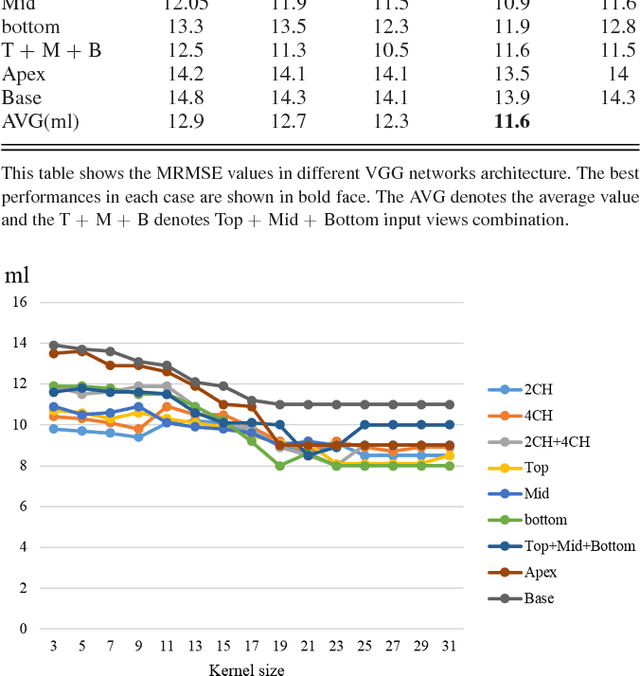
Abstract:Left ventricular (LV) volumes estimation is a critical procedure for cardiac disease diagnosis. The objective of this paper is to address direct LV volumes prediction task. Methods: In this paper, we propose a direct volumes prediction method based on the end-to-end deep convolutional neural networks (CNN). We study the end-to-end LV volumes prediction method in items of the data preprocessing, networks structure, and multi-views fusion strategy. The main contributions of this paper are the following aspects. First, we propose a new data preprocessing method on cardiac magnetic resonance (CMR). Second, we propose a new networks structure for end-to-end LV volumes estimation. Third, we explore the representational capacity of different slices, and propose a fusion strategy to improve the prediction accuracy. Results: The evaluation results show that the proposed method outperforms other state-of-the-art LV volumes estimation methods on the open accessible benchmark datasets. The clinical indexes derived from the predicted volumes agree well with the ground truth (EDV: R2=0.974, RMSE=9.6ml; ESV: R2=0.976, RMSE=7.1ml; EF: R2=0.828, RMSE =4.71%). Conclusion: Experimental results prove that the proposed method may be useful for LV volumes prediction task. Significance: The proposed method not only has application potential for cardiac diseases screening for large-scale CMR data, but also can be extended to other medical image research fields
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge