Ashley Mercado
VoxelAtlasGAN: 3D Left Ventricle Segmentation on Echocardiography with Atlas Guided Generation and Voxel-to-voxel Discrimination
Jun 10, 2018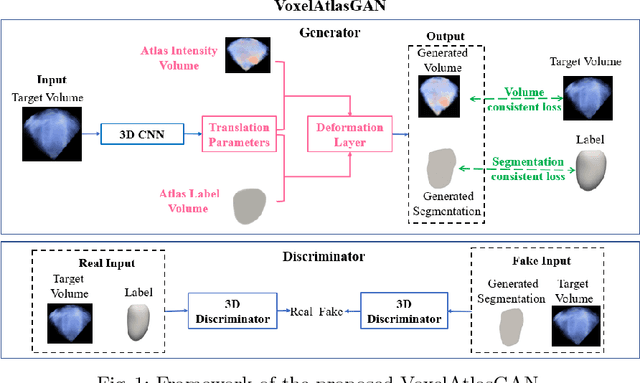

Abstract:3D left ventricle (LV) segmentation on echocardiography is very important for diagnosis and treatment of cardiac disease. It is not only because of that echocardiography is a real-time imaging technology and widespread in clinical application, but also because of that LV segmentation on 3D echocardiography can provide more full volume information of heart than LV segmentation on 2D echocardiography. However, 3D LV segmentation on echocardiography is still an open and challenging task owing to the lower contrast, higher noise and data dimensionality, limited annotation of 3D echocardiography. In this paper, we proposed a novel real-time framework, i.e., VoxelAtlasGAN, for 3D LV segmentation on 3D echocardiography. This framework has three contributions: 1) It is based on voxel-to-voxel conditional generative adversarial nets (cGAN). For the first time, cGAN is used for 3D LV segmentation on echocardiography. And cGAN advantageously fuses substantial 3D spatial context information from 3D echocardiography by self-learning structured loss; 2) For the first time, it embeds the atlas into an end-to-end optimization framework, which uses 3D LV atlas as a powerful prior knowledge to improve the inference speed, address the lower contrast and the limited annotation problems of 3D echocardiography; 3) It combines traditional discrimination loss and the new proposed consistent constraint, which further improves the generalization of the proposed framework. VoxelAtlasGAN was validated on 60 subjects on 3D echocardiography and it achieved satisfactory segmentation results and high inference speed. The mean surface distance is 1.85 mm, the mean hausdorff surface distance is 7.26 mm, mean dice is 0.953, the correlation of EF is 0.918, and the mean inference speed is 0.1s. These results have demonstrated that our proposed method has great potential for clinical application
Full Quantification of Left Ventricle via Deep Multitask Learning Network Respecting Intra- and Inter-Task Relatedness
Jun 14, 2017


Abstract:Cardiac left ventricle (LV) quantification is among the most clinically important tasks for identification and diagnosis of cardiac diseases, yet still a challenge due to the high variability of cardiac structure and the complexity of temporal dynamics. Full quantification, i.e., to simultaneously quantify all LV indices including two areas (cavity and myocardium), six regional wall thicknesses (RWT), three LV dimensions, and one cardiac phase, is even more challenging since the uncertain relatedness intra and inter each type of indices may hinder the learning procedure from better convergence and generalization. In this paper, we propose a newly-designed multitask learning network (FullLVNet), which is constituted by a deep convolution neural network (CNN) for expressive feature embedding of cardiac structure; two followed parallel recurrent neural network (RNN) modules for temporal dynamic modeling; and four linear models for the final estimation. During the final estimation, both intra- and inter-task relatedness are modeled to enforce improvement of generalization: 1) respecting intra-task relatedness, group lasso is applied to each of the regression tasks for sparse and common feature selection and consistent prediction; 2) respecting inter-task relatedness, three phase-guided constraints are proposed to penalize violation of the temporal behavior of the obtained LV indices. Experiments on MR sequences of 145 subjects show that FullLVNet achieves high accurate prediction with our intra- and inter-task relatedness, leading to MAE of 190mm$^2$, 1.41mm, 2.68mm for average areas, RWT, dimensions and error rate of 10.4\% for the phase classification. This endows our method a great potential in comprehensive clinical assessment of global, regional and dynamic cardiac function.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge