Yashu Liu
Advancing vision-language models in front-end development via data synthesis
Mar 03, 2025Abstract:Modern front-end (FE) development, especially when leveraging the unique features of frameworks like React and Vue, presents distinctive challenges. These include managing modular architectures, ensuring synchronization between data and visual outputs for declarative rendering, and adapting reusable components to various scenarios. Such complexities make it particularly difficult for state-of-the-art large vision-language models (VLMs) to generate accurate and functional code directly from design images. To address these challenges, we propose a reflective agentic workflow that synthesizes high-quality image-text data to capture the diverse characteristics of FE development. This workflow automates the extraction of self-contained\footnote{A \textbf{self-contained} code snippet is one that encapsulates all necessary logic, styling, and dependencies, ensuring it functions independently without requiring external imports or context.} code snippets from real-world projects, renders the corresponding visual outputs, and generates detailed descriptions that link design elements to functional code. To further expand the scope and utility of the synthesis, we introduce three data synthesis strategies: Evolution-based synthesis, which enables scalable and diverse dataset expansion; Waterfall-Model-based synthesis, which generates logically coherent code derived from system requirements; and Additive Development synthesis, which iteratively increases the complexity of human-authored components. We build a large vision-language model, Flame, trained on the synthesized datasets and demonstrate its effectiveness in generating React code via the $\text{pass}@k$ metric. Our results suggest that a code VLM trained to interpret images before code generation may achieve better performance.
Unsupervised Decomposition Networks for Bias Field Correction in MR Image
Jul 30, 2023



Abstract:Bias field, which is caused by imperfect MR devices or imaged objects, introduces intensity inhomogeneity into MR images and degrades the performance of MR image analysis methods. Many retrospective algorithms were developed to facilitate the bias correction, to which the deep learning-based methods outperformed. However, in the training phase, the supervised deep learning-based methods heavily rely on the synthesized bias field. As the formation of the bias field is extremely complex, it is difficult to mimic the true physical property of MR images by synthesized data. While bias field correction and image segmentation are strongly related, the segmentation map is precisely obtained by decoupling the bias field from the original MR image, and the bias value is indicated by the segmentation map in reverse. Thus, we proposed novel unsupervised decomposition networks that are trained only with biased data to obtain the bias-free MR images. Networks are made up of: a segmentation part to predict the probability of every pixel belonging to each class, and an estimation part to calculate the bias field, which are optimized alternately. Furthermore, loss functions based on the combination of fuzzy clustering and the multiplicative bias field are also devised. The proposed loss functions introduce the smoothness of bias field and construct the soft relationships among different classes under intra-consistency constraints. Extensive experiments demonstrate that the proposed method can accurately estimate bias fields and produce better bias correction results. The code is available on the link: https://github.com/LeongDong/Bias-Decomposition-Networks.
Cardiac Segmentation on Late Gadolinium Enhancement MRI: A Benchmark Study from Multi-Sequence Cardiac MR Segmentation Challenge
Jun 22, 2020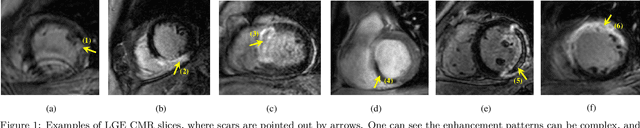
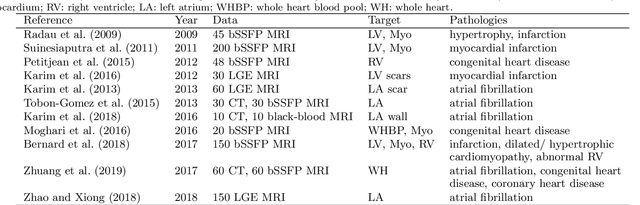
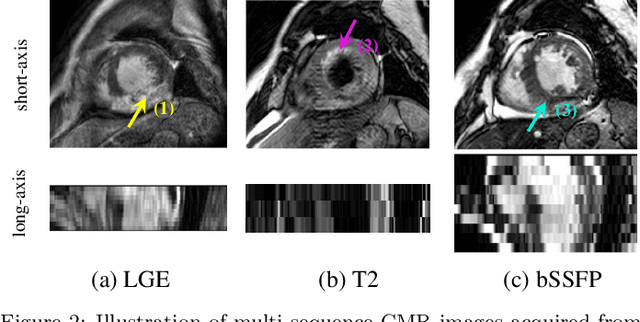
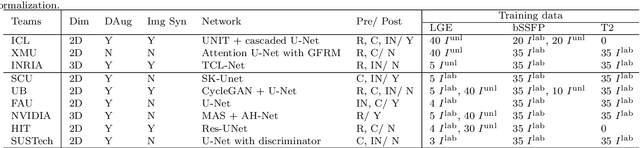
Abstract:Accurate computing, analysis and modeling of the ventricles and myocardium from medical images are important, especially in the diagnosis and treatment management for patients suffering from myocardial infarction (MI). Late gadolinium enhancement (LGE) cardiac magnetic resonance (CMR) provides an important protocol to visualize MI. However, automated segmentation of LGE CMR is still challenging, due to the indistinguishable boundaries, heterogeneous intensity distribution and complex enhancement patterns of pathological myocardium from LGE CMR. Furthermore, compared with the other sequences LGE CMR images with gold standard labels are particularly limited, which represents another obstacle for developing novel algorithms for automatic segmentation of LGE CMR. This paper presents the selective results from the Multi-Sequence Cardiac MR (MS-CMR) Segmentation challenge, in conjunction with MICCAI 2019. The challenge offered a data set of paired MS-CMR images, including auxiliary CMR sequences as well as LGE CMR, from 45 patients who underwent cardiomyopathy. It was aimed to develop new algorithms, as well as benchmark existing ones for LGE CMR segmentation and compare them objectively. In addition, the paired MS-CMR images could enable algorithms to combine the complementary information from the other sequences for the segmentation of LGE CMR. Nine representative works were selected for evaluation and comparisons, among which three methods are unsupervised methods and the other six are supervised. The results showed that the average performance of the nine methods was comparable to the inter-observer variations. The success of these methods was mainly attributed to the inclusion of the auxiliary sequences from the MS-CMR images, which provide important label information for the training of deep neural networks.
A Global Benchmark of Algorithms for Segmenting Late Gadolinium-Enhanced Cardiac Magnetic Resonance Imaging
May 07, 2020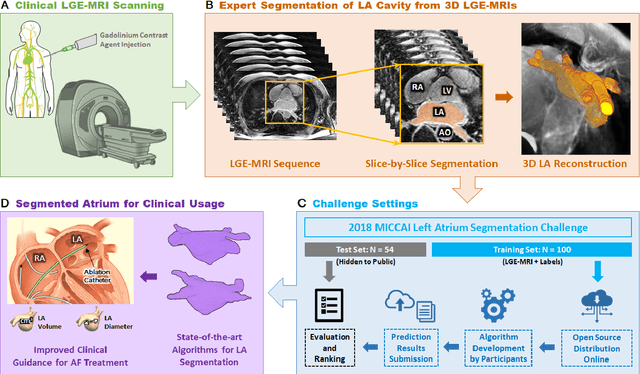
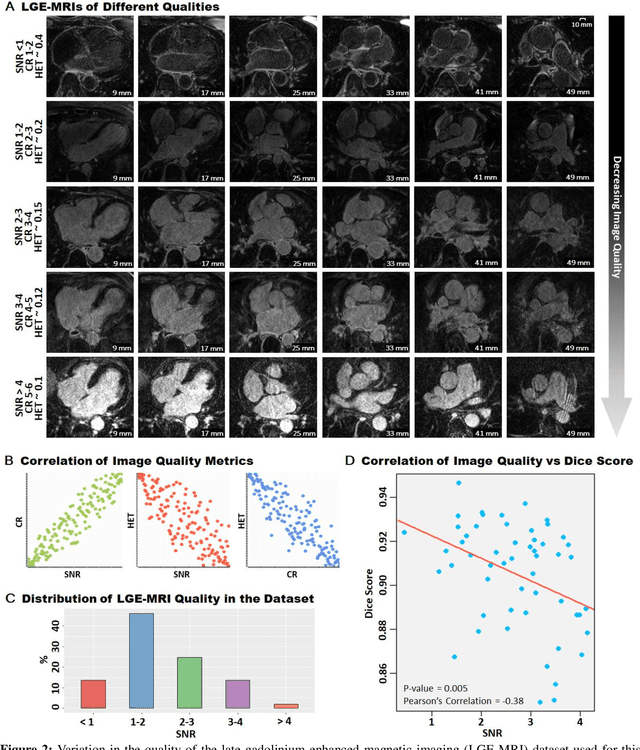
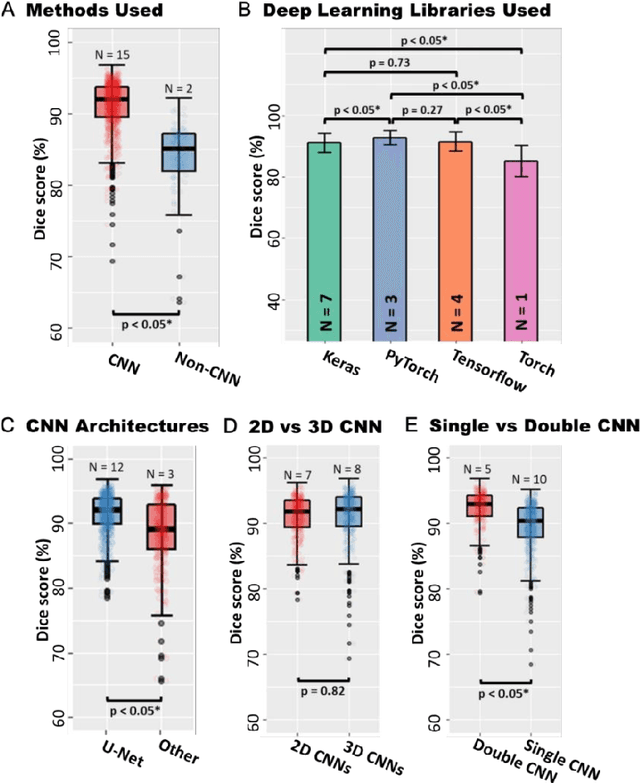
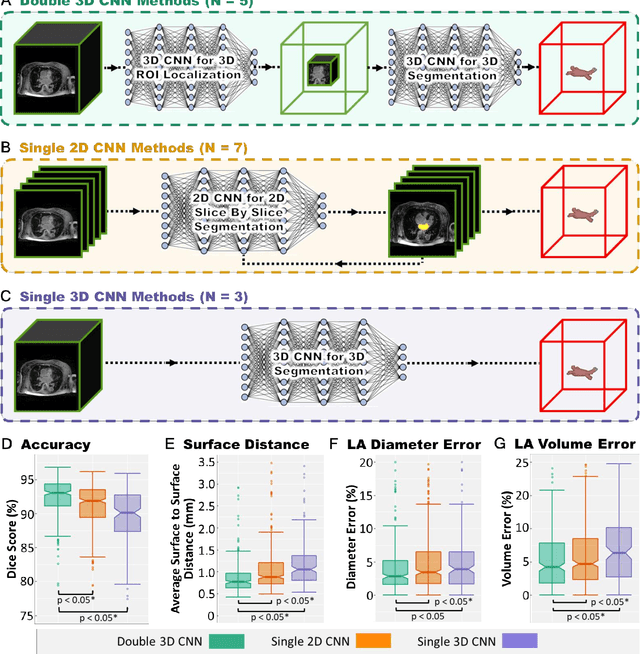
Abstract:Segmentation of cardiac images, particularly late gadolinium-enhanced magnetic resonance imaging (LGE-MRI) widely used for visualizing diseased cardiac structures, is a crucial first step for clinical diagnosis and treatment. However, direct segmentation of LGE-MRIs is challenging due to its attenuated contrast. Since most clinical studies have relied on manual and labor-intensive approaches, automatic methods are of high interest, particularly optimized machine learning approaches. To address this, we organized the "2018 Left Atrium Segmentation Challenge" using 154 3D LGE-MRIs, currently the world's largest cardiac LGE-MRI dataset, and associated labels of the left atrium segmented by three medical experts, ultimately attracting the participation of 27 international teams. In this paper, extensive analysis of the submitted algorithms using technical and biological metrics was performed by undergoing subgroup analysis and conducting hyper-parameter analysis, offering an overall picture of the major design choices of convolutional neural networks (CNNs) and practical considerations for achieving state-of-the-art left atrium segmentation. Results show the top method achieved a dice score of 93.2% and a mean surface to a surface distance of 0.7 mm, significantly outperforming prior state-of-the-art. Particularly, our analysis demonstrated that double, sequentially used CNNs, in which a first CNN is used for automatic region-of-interest localization and a subsequent CNN is used for refined regional segmentation, achieved far superior results than traditional methods and pipelines containing single CNNs. This large-scale benchmarking study makes a significant step towards much-improved segmentation methods for cardiac LGE-MRIs, and will serve as an important benchmark for evaluating and comparing the future works in the field.
An Automatic Cardiac Segmentation Framework based on Multi-sequence MR Image
Sep 12, 2019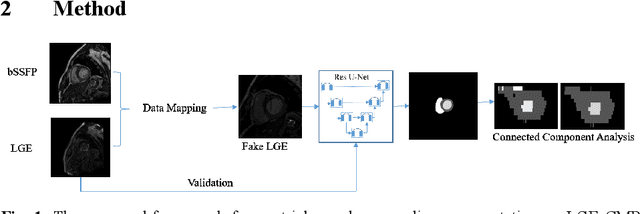
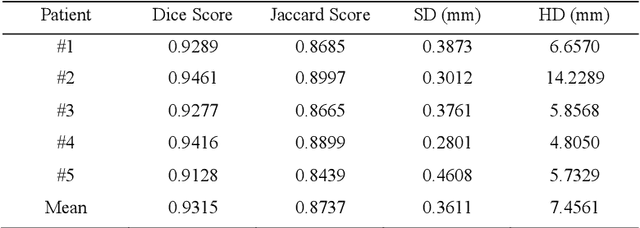
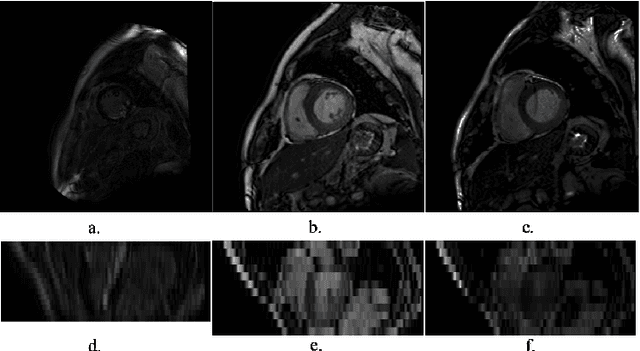
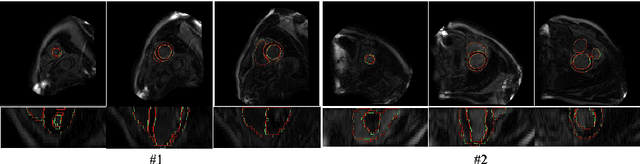
Abstract:LGE CMR is an efficient technology for detecting infarcted myocardium. An efficient and objective ventricle segmentation method in LGE can benefit the location of the infarcted myocardium. In this paper, we proposed an automatic framework for LGE image segmentation. There are just 5 labeled LGE volumes with about 15 slices of each volume. We adopted histogram match, an invariant of rotation registration method, on the other labeled modalities to achieve effective augmentation of the training data. A CNN segmentation model was trained based on the augmented training data by leave-one-out strategy. The predicted result of the model followed a connected component analysis for each class to remain the largest connected component as the final segmentation result. Our model was evaluated by the 2019 Multi-sequence Cardiac MR Segmentation Challenge. The mean testing result of 40 testing volumes on Dice score, Jaccard score, Surface distance, and Hausdorff distance is 0.8087, 0.6976, 2.8727mm, and 15.6387mm, respectively. The experiment result shows a satisfying performance of the proposed framework. Code is available at https://github.com/Suiiyu/MS-CMR2019.
* accepted by STACOM 2019
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge