Qian Tao
Enabling Ultra-Fast Cardiovascular Imaging Across Heterogeneous Clinical Environments with a Generalist Foundation Model and Multimodal Database
Dec 25, 2025Abstract:Multimodal cardiovascular magnetic resonance (CMR) imaging provides comprehensive and non-invasive insights into cardiovascular disease (CVD) diagnosis and underlying mechanisms. Despite decades of advancements, its widespread clinical adoption remains constrained by prolonged scan times and heterogeneity across medical environments. This underscores the urgent need for a generalist reconstruction foundation model for ultra-fast CMR imaging, one capable of adapting across diverse imaging scenarios and serving as the essential substrate for all downstream analyses. To enable this goal, we curate MMCMR-427K, the largest and most comprehensive multimodal CMR k-space database to date, comprising 427,465 multi-coil k-space data paired with structured metadata across 13 international centers, 12 CMR modalities, 15 scanners, and 17 CVD categories in populations across three continents. Building on this unprecedented resource, we introduce CardioMM, a generalist reconstruction foundation model capable of dynamically adapting to heterogeneous fast CMR imaging scenarios. CardioMM unifies semantic contextual understanding with physics-informed data consistency to deliver robust reconstructions across varied scanners, protocols, and patient presentations. Comprehensive evaluations demonstrate that CardioMM achieves state-of-the-art performance in the internal centers and exhibits strong zero-shot generalization to unseen external settings. Even at imaging acceleration up to 24x, CardioMM reliably preserves key cardiac phenotypes, quantitative myocardial biomarkers, and diagnostic image quality, enabling a substantial increase in CMR examination throughput without compromising clinical integrity. Together, our open-access MMCMR-427K database and CardioMM framework establish a scalable pathway toward high-throughput, high-quality, and clinically accessible cardiovascular imaging.
Groupwise Registration with Physics-Informed Test-Time Adaptation on Multi-parametric Cardiac MRI
Oct 29, 2025Abstract:Multiparametric mapping MRI has become a viable tool for myocardial tissue characterization. However, misalignment between multiparametric maps makes pixel-wise analysis challenging. To address this challenge, we developed a generalizable physics-informed deep-learning model using test-time adaptation to enable group image registration across contrast weighted images acquired from multiple physical models (e.g., a T1 mapping model and T2 mapping model). The physics-informed adaptation utilized the synthetic images from specific physics model as registration reference, allows for transductive learning for various tissue contrast. We validated the model in healthy volunteers with various MRI sequences, demonstrating its improvement for multi-modal registration with a wide range of image contrast variability.
Beyond the LUMIR challenge: The pathway to foundational registration models
May 30, 2025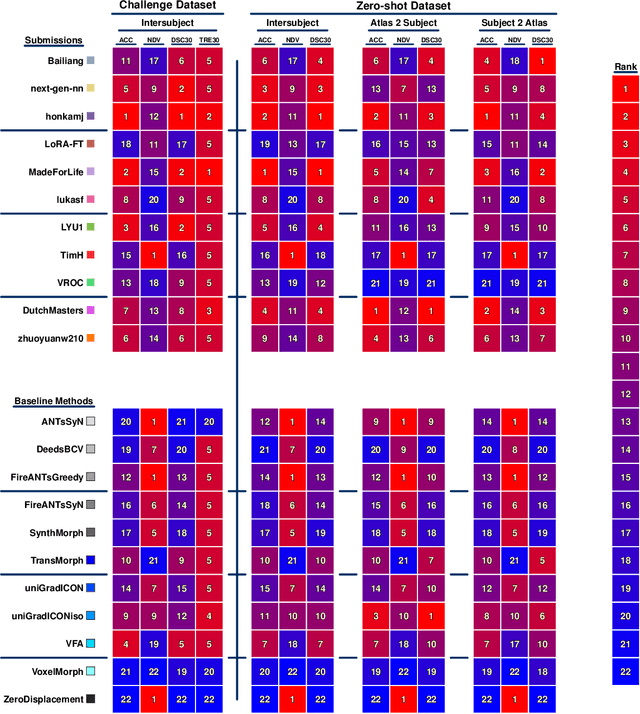
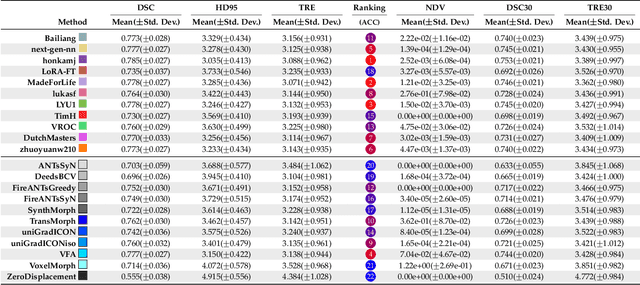
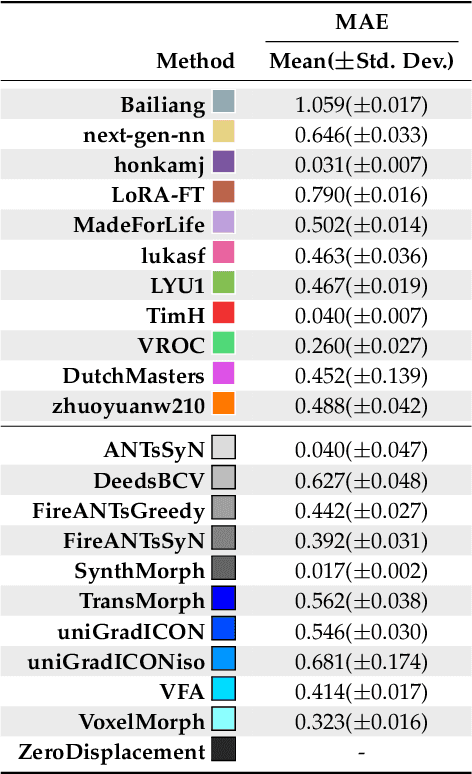
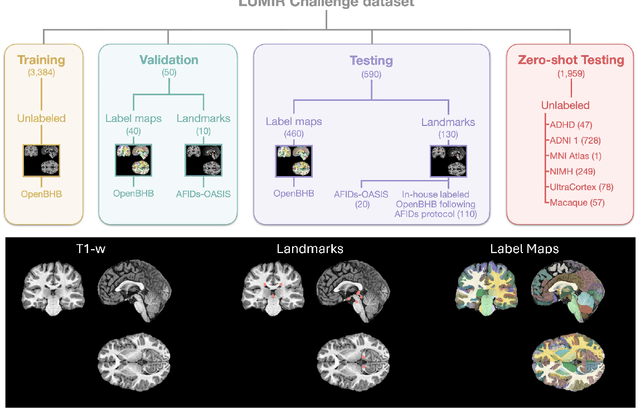
Abstract:Medical image challenges have played a transformative role in advancing the field, catalyzing algorithmic innovation and establishing new performance standards across diverse clinical applications. Image registration, a foundational task in neuroimaging pipelines, has similarly benefited from the Learn2Reg initiative. Building on this foundation, we introduce the Large-scale Unsupervised Brain MRI Image Registration (LUMIR) challenge, a next-generation benchmark designed to assess and advance unsupervised brain MRI registration. Distinct from prior challenges that leveraged anatomical label maps for supervision, LUMIR removes this dependency by providing over 4,000 preprocessed T1-weighted brain MRIs for training without any label maps, encouraging biologically plausible deformation modeling through self-supervision. In addition to evaluating performance on 590 held-out test subjects, LUMIR introduces a rigorous suite of zero-shot generalization tasks, spanning out-of-domain imaging modalities (e.g., FLAIR, T2-weighted, T2*-weighted), disease populations (e.g., Alzheimer's disease), acquisition protocols (e.g., 9.4T MRI), and species (e.g., macaque brains). A total of 1,158 subjects and over 4,000 image pairs were included for evaluation. Performance was assessed using both segmentation-based metrics (Dice coefficient, 95th percentile Hausdorff distance) and landmark-based registration accuracy (target registration error). Across both in-domain and zero-shot tasks, deep learning-based methods consistently achieved state-of-the-art accuracy while producing anatomically plausible deformation fields. The top-performing deep learning-based models demonstrated diffeomorphic properties and inverse consistency, outperforming several leading optimization-based methods, and showing strong robustness to most domain shifts, the exception being a drop in performance on out-of-domain contrasts.
Towards Universal Learning-based Model for Cardiac Image Reconstruction: Summary of the CMRxRecon2024 Challenge
Mar 05, 2025Abstract:Cardiovascular magnetic resonance (CMR) offers diverse imaging contrasts for assessment of cardiac function and tissue characterization. However, acquiring each single CMR modality is often time-consuming, and comprehensive clinical protocols require multiple modalities with various sampling patterns, further extending the overall acquisition time and increasing susceptibility to motion artifacts. Existing deep learning-based reconstruction methods are often designed for specific acquisition parameters, which limits their ability to generalize across a variety of scan scenarios. As part of the CMRxRecon Series, the CMRxRecon2024 challenge provides diverse datasets encompassing multi-modality multi-view imaging with various sampling patterns, and a platform for the international community to develop and benchmark reconstruction solutions in two well-crafted tasks. Task 1 is a modality-universal setting, evaluating the out-of-distribution generalization of the reconstructed model, while Task 2 follows sampling-universal setting assessing the one-for-all adaptability of the universal model. Main contributions include providing the first and largest publicly available multi-modality, multi-view cardiac k-space dataset; developing a benchmarking platform that simulates clinical acceleration protocols, with a shared code library and tutorial for various k-t undersampling patterns and data processing; giving technical insights of enhanced data consistency based on physic-informed networks and adaptive prompt-learning embedding to be versatile to different clinical settings; additional finding on evaluation metrics to address the limitations of conventional ground-truth references in universal reconstruction tasks.
Exact Acceleration of Subgraph Graph Neural Networks by Eliminating Computation Redundancy
Dec 24, 2024



Abstract:Graph neural networks (GNNs) have become a prevalent framework for graph tasks. Many recent studies have proposed the use of graph convolution methods over the numerous subgraphs of each graph, a concept known as subgraph graph neural networks (subgraph GNNs), to enhance GNNs' ability to distinguish non-isomorphic graphs. To maximize the expressiveness, subgraph GNNs often require each subgraph to have equal size to the original graph. Despite their impressive performance, subgraph GNNs face challenges due to the vast number and large size of subgraphs which lead to a surge in training data, resulting in both storage and computational inefficiencies. In response to this problem, this paper introduces Ego-Nets-Fit-All (ENFA), a model that uniformly takes the smaller ego nets as subgraphs, thereby providing greater storage and computational efficiency, while at the same time guarantees identical outputs to the original subgraph GNNs even taking the whole graph as subgraphs. The key is to identify and eliminate the redundant computation among subgraphs. For example, a node $v_i$ may appear in multiple subgraphs but is far away from all of their centers (the unsymmetric part between subgraphs). Therefore, its first few rounds of message passing within each subgraph can be computed once in the original graph instead of being computed multiple times within each subgraph. Such strategy enables our ENFA to accelerate subgraph GNNs in an exact way, unlike previous sampling approaches that often lose the performance. Extensive experiments across various datasets reveal that compared with the conventional subgraph GNNs, ENFA can reduce storage space by 29.0% to 84.5% and improve training efficiency by up to 1.66x.
GL-Fusion: Rethinking the Combination of Graph Neural Network and Large Language model
Dec 08, 2024



Abstract:Recent research on integrating Large Language Models (LLMs) with Graph Neural Networks (GNNs) typically follows two approaches: LLM-centered models, which convert graph data into tokens for LLM processing, and GNN-centered models, which use LLMs to encode text features into node and edge representations for GNN input. LLM-centered models often struggle to capture graph structures effectively, while GNN-centered models compress variable-length textual data into fixed-size vectors, limiting their ability to understand complex semantics. Additionally, GNN-centered approaches require converting tasks into a uniform, manually-designed format, restricting them to classification tasks and preventing language output. To address these limitations, we introduce a new architecture that deeply integrates GNN with LLM, featuring three key innovations: (1) Structure-Aware Transformers, which incorporate GNN's message-passing capabilities directly into LLM's transformer layers, allowing simultaneous processing of textual and structural information and generating outputs from both GNN and LLM; (2) Graph-Text Cross-Attention, which processes full, uncompressed text from graph nodes and edges, ensuring complete semantic integration; and (3) GNN-LLM Twin Predictor, enabling LLM's flexible autoregressive generation alongside GNN's scalable one-pass prediction. GL-Fusion achieves outstand performance on various tasks. Notably, it achieves state-of-the-art performance on OGBN-Arxiv and OGBG-Code2.
Improving Instance Optimization in Deformable Image Registration with Gradient Projection
Oct 21, 2024


Abstract:Deformable image registration is inherently a multi-objective optimization (MOO) problem, requiring a delicate balance between image similarity and deformation regularity. These conflicting objectives often lead to poor optimization outcomes, such as being trapped in unsatisfactory local minima or experiencing slow convergence. Deep learning methods have recently gained popularity in this domain due to their efficiency in processing large datasets and achieving high accuracy. However, they often underperform during test time compared to traditional optimization techniques, which further explore iterative, instance-specific gradient-based optimization. This performance gap is more pronounced when a distribution shift between training and test data exists. To address this issue, we focus on the instance optimization (IO) paradigm, which involves additional optimization for test-time instances based on a pre-trained model. IO effectively combines the generalization capabilities of deep learning with the fine-tuning advantages of instance-specific optimization. Within this framework, we emphasize the use of gradient projection to mitigate conflicting updates in MOO. This technique projects conflicting gradients into a common space, better aligning the dual objectives and enhancing optimization stability. We validate our method using a state-of-the-art foundation model on the 3D Brain inter-subject registration task (LUMIR) from the Learn2Reg 2024 Challenge. Our results show significant improvements over standard gradient descent, leading to more accurate and reliable registration results.
AsymKV: Enabling 1-Bit Quantization of KV Cache with Layer-Wise Asymmetric Quantization Configurations
Oct 17, 2024



Abstract:Large language models have shown exceptional capabilities in a wide range of tasks, such as text generation and video generation, among others. However, due to their massive parameter count, these models often require substantial storage space, imposing significant constraints on the machines deploying LLMs. To overcome this limitation, one research direction proposes to compress the models using integer replacements for floating-point numbers, in a process known as Quantization. Some recent studies suggest quantizing the key and value cache (KV Cache) of LLMs, and designing quantization techniques that treat the key and value matrices equivalently. This work delves deeper into the asymmetric structural roles of KV Cache, a phenomenon where the transformer's output loss is more sensitive to the quantization of key matrices. We conduct a systematic examination of the attention output error resulting from key and value quantization. The phenomenon inspires us to propose an asymmetric quantization strategy. Our approach allows for 1-bit quantization of the KV cache by implementing distinct configurations for key and value matrices. We carry out experiments across a variety of datasets, demonstrating that our proposed model allows for the quantization of up to 75% decoder layers with 1 bit, while simultaneously maintaining performance levels comparable to those of the models with floating parameters.
Lost in Tracking: Uncertainty-guided Cardiac Cine MRI Segmentation at Right Ventricle Base
Oct 04, 2024Abstract:Accurate biventricular segmentation of cardiac magnetic resonance (CMR) cine images is essential for the clinical evaluation of heart function. However, compared to left ventricle (LV), right ventricle (RV) segmentation is still more challenging and less reproducible. Degenerate performance frequently occurs at the RV base, where the in-plane anatomical structures are complex (with atria, valve, and aorta) and vary due to the strong interplanar motion. In this work, we propose to address the currently unsolved issues in CMR segmentation, specifically at the RV base, with two strategies: first, we complemented the public resource by reannotating the RV base in the ACDC dataset, with refined delineation of the right ventricle outflow tract (RVOT), under the guidance of an expert cardiologist. Second, we proposed a novel dual encoder U-Net architecture that leverages temporal incoherence to inform the segmentation when interplanar motions occur. The inter-planar motion is characterized by loss-of-tracking, via Bayesian uncertainty of a motion-tracking model. Our experiments showed that our method significantly improved RV base segmentation taking into account temporal incoherence. Furthermore, we investigated the reproducibility of deep learning-based segmentation and showed that the combination of consistent annotation and loss of tracking could enhance the reproducibility of RV segmentation, potentially facilitating a large number of clinical studies focusing on RV.
Deep-learning-based groupwise registration for motion correction of cardiac $T_1$ mapping
Jun 21, 2024Abstract:Quantitative $T_1$ mapping by MRI is an increasingly important tool for clinical assessment of cardiovascular diseases. The cardiac $T_1$ map is derived by fitting a known signal model to a series of baseline images, while the quality of this map can be deteriorated by involuntary respiratory and cardiac motion. To correct motion, a template image is often needed to register all baseline images, but the choice of template is nontrivial, leading to inconsistent performance sensitive to image contrast. In this work, we propose a novel deep-learning-based groupwise registration framework, which omits the need for a template, and registers all baseline images simultaneously. We design two groupwise losses for this registration framework: the first is a linear principal component analysis (PCA) loss that enforces alignment of baseline images irrespective of the intensity variation, and the second is an auxiliary relaxometry loss that enforces adherence of intensity profile to the signal model. We extensively evaluated our method, termed ``PCA-Relax'', and other baseline methods on an in-house cardiac MRI dataset including both pre- and post-contrast $T_1$ sequences. All methods were evaluated under three distinct training-and-evaluation strategies, namely, standard, one-shot, and test-time-adaptation. The proposed PCA-Relax showed further improved performance of registration and mapping over well-established baselines. The proposed groupwise framework is generic and can be adapted to applications involving multiple images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge