Peter Kellman
Imaging Transformer for MRI Denoising: a Scalable Model Architecture that enables SNR << 1 Imaging
Apr 13, 2025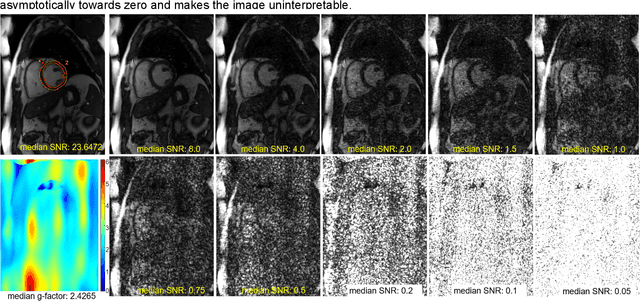

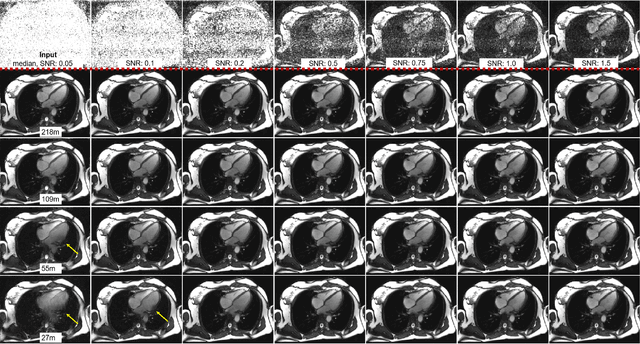
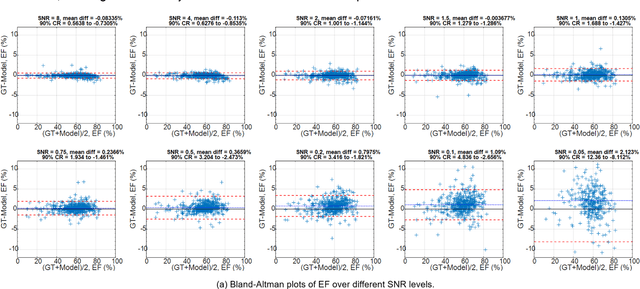
Abstract:Purpose: To propose a flexible and scalable imaging transformer (IT) architecture with three attention modules for multi-dimensional imaging data and apply it to MRI denoising with very low input SNR. Methods: Three independent attention modules were developed: spatial local, spatial global, and frame attentions. They capture long-range signal correlation and bring back the locality of information in images. An attention-cell-block design processes 5D tensors ([B, C, F, H, W]) for 2D, 2D+T, and 3D image data. A High Resolution (HRNet) backbone was built to hold IT blocks. Training dataset consists of 206,677 cine series and test datasets had 7,267 series. Ten input SNR levels from 0.05 to 8.0 were tested. IT models were compared to seven convolutional and transformer baselines. To test scalability, four IT models 27m to 218m parameters were trained. Two senior cardiologists reviewed IT model outputs from which the EF was measured and compared against the ground-truth. Results: IT models significantly outperformed other models over the tested SNR levels. The performance gap was most prominent at low SNR levels. The IT-218m model had the highest SSIM and PSNR, restoring good image quality and anatomical details even at SNR 0.2. Two experts agreed at this SNR or above, the IT model output gave the same clinical interpretation as the ground-truth. The model produced images that had accurate EF measurements compared to ground-truth values. Conclusions: Imaging transformer model offers strong performance, scalability, and versatility for MR denoising. It recovers image quality suitable for confident clinical reading and accurate EF measurement, even at very low input SNR of 0.2.
Recurrent Inference Machine for Medical Image Registration
Jun 19, 2024Abstract:Image registration is essential for medical image applications where alignment of voxels across multiple images is needed for qualitative or quantitative analysis. With recent advancements in deep neural networks and parallel computing, deep learning-based medical image registration methods become competitive with their flexible modelling and fast inference capabilities. However, compared to traditional optimization-based registration methods, the speed advantage may come at the cost of registration performance at inference time. Besides, deep neural networks ideally demand large training datasets while optimization-based methods are training-free. To improve registration accuracy and data efficiency, we propose a novel image registration method, termed Recurrent Inference Image Registration (RIIR) network. RIIR is formulated as a meta-learning solver to the registration problem in an iterative manner. RIIR addresses the accuracy and data efficiency issues, by learning the update rule of optimization, with implicit regularization combined with explicit gradient input. We evaluated RIIR extensively on brain MRI and quantitative cardiac MRI datasets, in terms of both registration accuracy and training data efficiency. Our experiments showed that RIIR outperformed a range of deep learning-based methods, even with only $5\%$ of the training data, demonstrating high data efficiency. Key findings from our ablation studies highlighted the important added value of the hidden states introduced in the recurrent inference framework for meta-learning. Our proposed RIIR offers a highly data-efficient framework for deep learning-based medical image registration.
Inline AI: Open-source Deep Learning Inference for Cardiac MR
Apr 03, 2024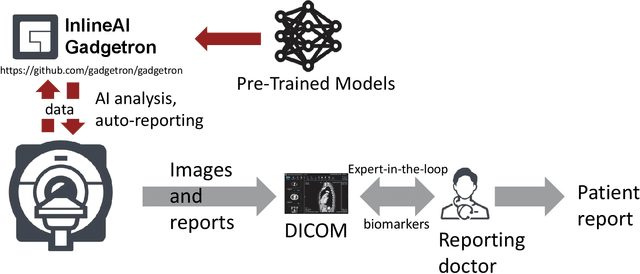
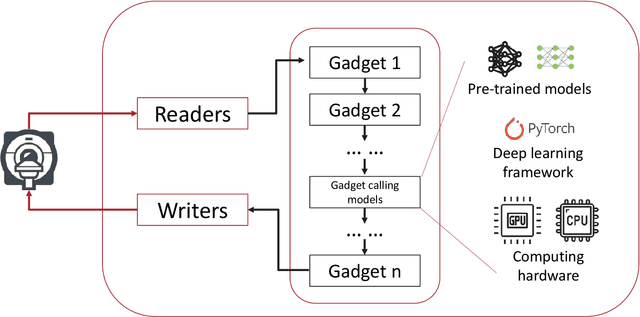
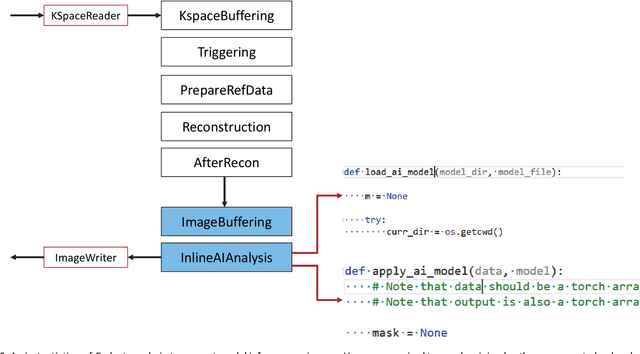
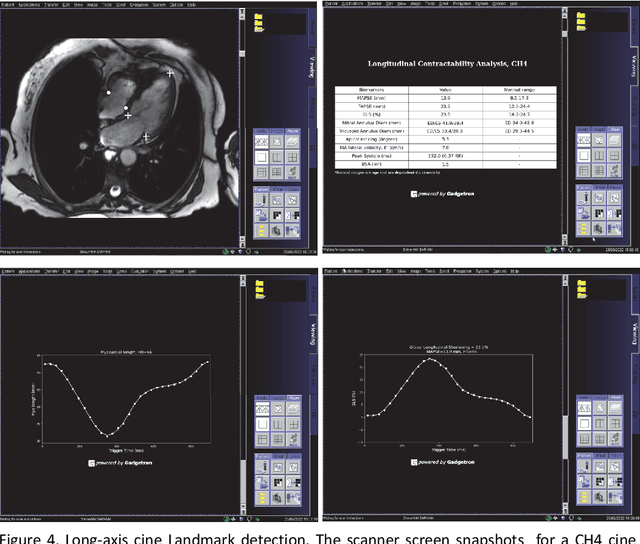
Abstract:Cardiac Magnetic Resonance (CMR) is established as a non-invasive imaging technique for evaluation of heart function, anatomy, and myocardial tissue characterization. Quantitative biomarkers are central for diagnosis and management of heart disease. Deep learning (DL) is playing an ever more important role in extracting these quantitative measures from CMR images. While many researchers have reported promising results in training and evaluating models, model deployment into the imaging workflow is less explored. A new imaging AI framework, the InlineAI, was developed and open-sourced. The main innovation is to enable the model inference inline as a part of imaging computation, instead of as an offline post-processing step and to allow users to plug in their models. We demonstrate the system capability on three applications: long-axis CMR cine landmark detection, short-axis CMR cine analysis of function and anatomy, and quantitative perfusion mapping. The InlineAI allowed models to be deployed into imaging workflow in a streaming manner directly on the scanner. The model was loaded and inference on incoming images were performed while the data acquisition was ongoing, and results were sent back to scanner. Several biomarkers were extracted from model outputs in the demonstrated applications and reported as curves and tabular values. All processes are full automated. the model inference was completed within 6-45s after the end of imaging data acquisition.
Imaging transformer for MRI denoising with the SNR unit training: enabling generalization across field-strengths, imaging contrasts, and anatomy
Apr 03, 2024

Abstract:The ability to recover MRI signal from noise is key to achieve fast acquisition, accurate quantification, and high image quality. Past work has shown convolutional neural networks can be used with abundant and paired low and high-SNR images for training. However, for applications where high-SNR data is difficult to produce at scale (e.g. with aggressive acceleration, high resolution, or low field strength), training a new denoising network using a large quantity of high-SNR images can be infeasible. In this study, we overcome this limitation by improving the generalization of denoising models, enabling application to many settings beyond what appears in the training data. Specifically, we a) develop a training scheme that uses complex MRIs reconstructed in the SNR units (i.e., the images have a fixed noise level, SNR unit training) and augments images with realistic noise based on coil g-factor, and b) develop a novel imaging transformer (imformer) to handle 2D, 2D+T, and 3D MRIs in one model architecture. Through empirical evaluation, we show this combination improves performance compared to CNN models and improves generalization, enabling a denoising model to be used across field-strengths, image contrasts, and anatomy.
Landmark detection in Cardiac Magnetic Resonance Imaging Using A Convolutional Neural Network
Aug 14, 2020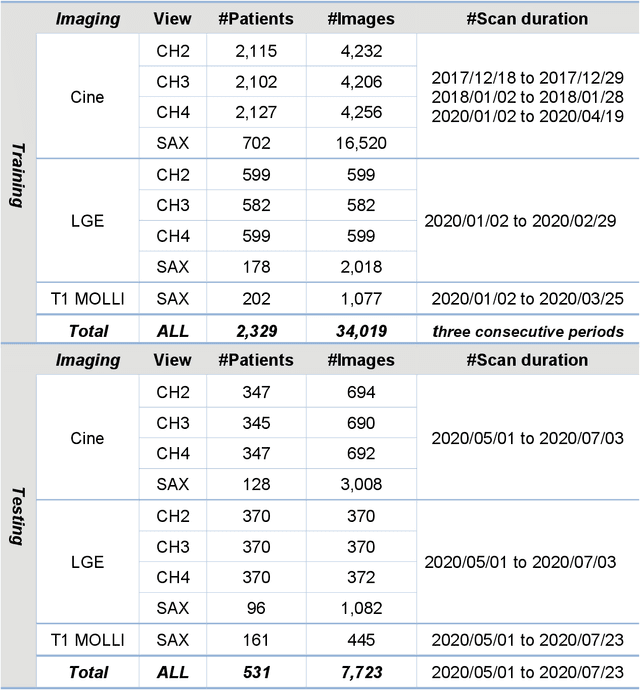
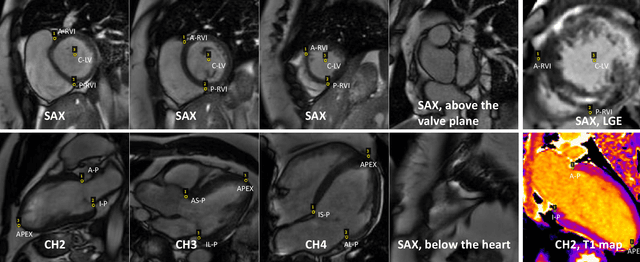
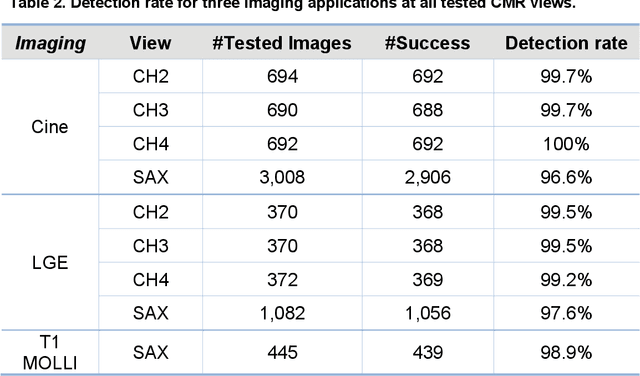
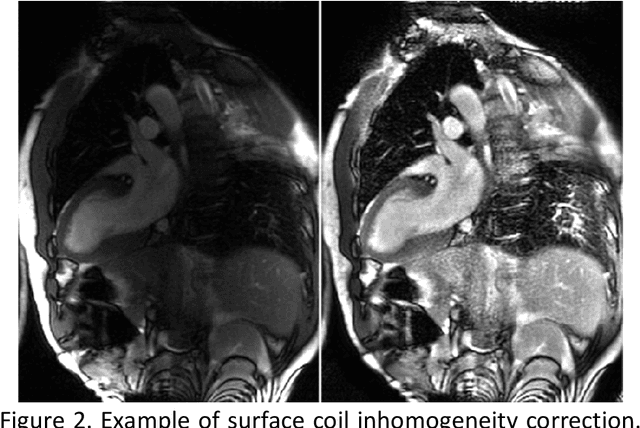
Abstract:Purpose: To develop a convolutional neural network (CNN) solution for robust landmark detection in cardiac MR images. Methods: This retrospective study included cine, LGE and T1 mapping scans from two hospitals. The training set included 2,329 patients and 34,019 images. A hold-out test set included 531 patients and 7,723 images. CNN models were developed to detect two mitral valve plane and apical points on long-axis (LAX) images. On short-axis (SAX) images, anterior and posterior RV insertion points and LV center were detected. Model outputs were compared to manual labels by two operators for accuracy with a t-test for statistical significance. The trained model was deployed to MR scanners. Results: For the LAX images, success detection was 99.8% for cine, 99.4% for LGE. For the SAX, success rate was 96.6%, 97.6% and 98.9% for cine, LGE and T1-mapping. The L2 distances between model and manual labels were 2 to 3.5 mm, indicating close agreement between model landmarks to manual labels. No significant differences were found for the anterior RV insertion angle and LV length by the models and operators for all views and imaging sequences. Model inference on MR scanner took 610ms/5.6s on GPU/CPU, respectively, for a typical cardiac cine series. Conclusions: This study developed, validated and deployed a CNN solution for robust landmark detection in both long and short-axis CMR images for cine, LGE and T1 mapping sequences, with the accuracy comparable to the inter-operator variation.
Automated Inline Analysis of Myocardial Perfusion MRI with Deep Learning
Nov 02, 2019



Abstract:Recent development of quantitative myocardial blood flow (MBF) mapping allows direct evaluation of absolute myocardial perfusion, by computing pixel-wise flow maps. Clinical studies suggest quantitative evaluation would be more desirable for objectivity and efficiency. Objective assessment can be further facilitated by segmenting the myocardium and automatically generating reports following the AHA model. This will free user interaction for analysis and lead to a 'one-click' solution to improve workflow. This paper proposes a deep neural network based computational workflow for inline myocardial perfusion analysis. Adenosine stress and rest perfusion scans were acquired from three hospitals. Training set included N=1,825 perfusion series from 1,034 patients. Independent test set included 200 scans from 105 patients. Data were consecutively acquired at each site. A convolution neural net (CNN) model was trained to provide segmentation for LV cavity, myocardium and right ventricular by processing incoming 2D+T perfusion Gd series. Model outputs were compared to manual ground-truth for accuracy of segmentation and flow measures derived on global and per-sector basis. The trained models were integrated onto MR scanners for effective inference. Segmentation accuracy and myocardial flow measures were compared between CNN models and manual ground-truth. The mean Dice ratio of CNN derived myocardium was 0.93 +/- 0.04. Both global flow and per-sector values showed no significant difference, compared to manual results. The AHA 16 segment model was automatically generated and reported on the MR scanner. As a result, the fully automated analysis of perfusion flow mapping was achieved. This solution was integrated on the MR scanner, enabling 'one-click' analysis and reporting of myocardial blood flow.
Automated Detection of Left Ventricle in Arterial Input Function Images for Inline Perfusion Mapping using Deep Learning: A study of 15,000 Patients
Oct 16, 2019


Abstract:Quantification of myocardial perfusion has the potential to improve detection of regional and global flow reduction. Significant effort has been made to automate the workflow, where one essential step is the arterial input function (AIF) extraction. Since failure here invalidates quantification, high accuracy is required. For this purpose, this study presents a robust AIF detection method using the convolutional neural net (CNN) model. CNN models were trained by assembling 25,027 scans (N=12,984 patients) from three hospitals, seven scanners. A test set of 5,721 scans (N=2,805 patients) evaluated model performance. The 2D+T AIF time series was inputted into CNN. Two variations were investigated: a) Two Classes (2CS) for background and foreground (LV mask); b) Three Classes (3CS) for background, foreground LV and RV. Final model was deployed on MR scanners via the Gadgetron InlineAI. Model loading time on MR scanner was ~340ms and applying it took ~180ms. The 3CS model successfully detect LV for 99.98% of all test cases (1 failed out of 5,721 cases). The mean Dice ratio for 3CS was 0.87+/-0.08 with 92.0% of all test cases having Dice ratio >0.75, while the 2CS model gave lower Dice of 0.82+/-0.22 (P<1e-5). Extracted AIF signals using CNN were further compared to manual ground-truth for foot-time, peak-time, first-pass duration, peak value and area-under-curve. No significant differences were found for all features (P>0.2). This study proposed, validated, and deployed a robust CNN solution to detect the LV for the extraction of the AIF signal used in fully automated perfusion flow mapping. A very large data cohort was assembled and resulting models were deployed to MR scanners for fully inline AI in clinical hospitals.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge