James Howard
Inline AI: Open-source Deep Learning Inference for Cardiac MR
Apr 03, 2024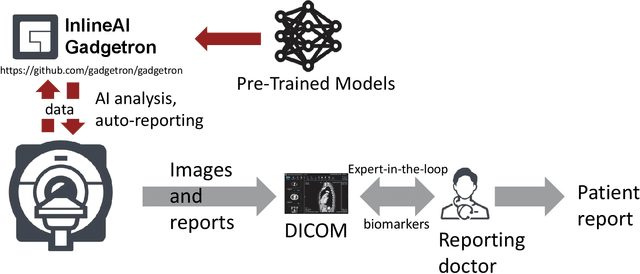
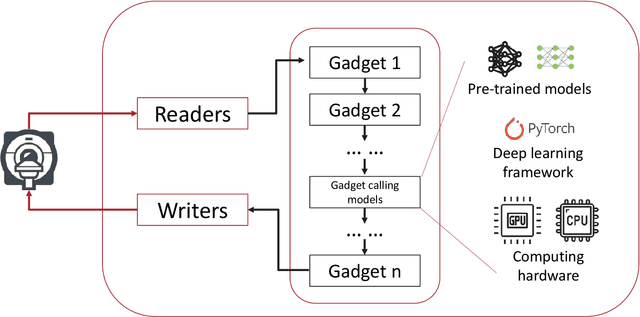
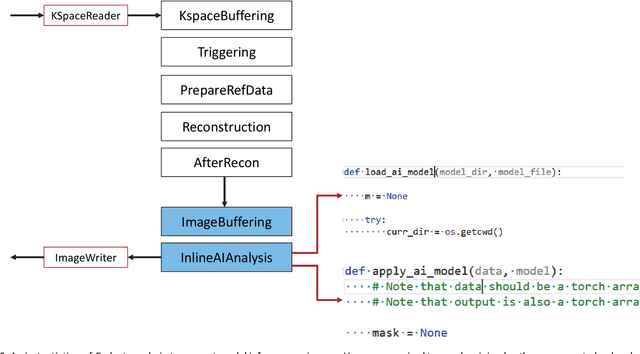
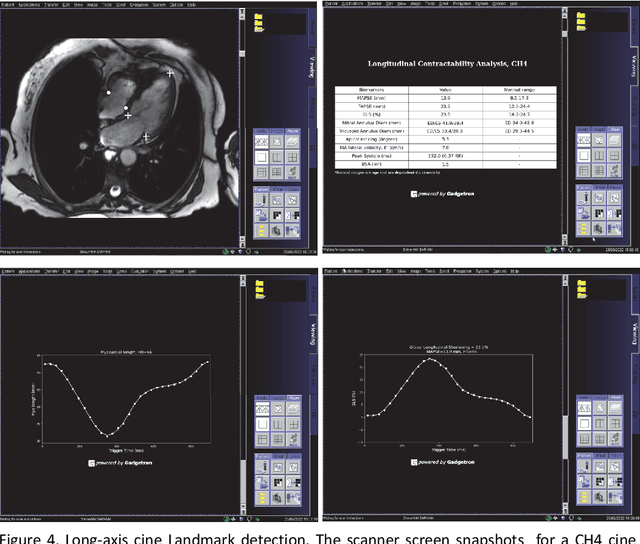
Abstract:Cardiac Magnetic Resonance (CMR) is established as a non-invasive imaging technique for evaluation of heart function, anatomy, and myocardial tissue characterization. Quantitative biomarkers are central for diagnosis and management of heart disease. Deep learning (DL) is playing an ever more important role in extracting these quantitative measures from CMR images. While many researchers have reported promising results in training and evaluating models, model deployment into the imaging workflow is less explored. A new imaging AI framework, the InlineAI, was developed and open-sourced. The main innovation is to enable the model inference inline as a part of imaging computation, instead of as an offline post-processing step and to allow users to plug in their models. We demonstrate the system capability on three applications: long-axis CMR cine landmark detection, short-axis CMR cine analysis of function and anatomy, and quantitative perfusion mapping. The InlineAI allowed models to be deployed into imaging workflow in a streaming manner directly on the scanner. The model was loaded and inference on incoming images were performed while the data acquisition was ongoing, and results were sent back to scanner. Several biomarkers were extracted from model outputs in the demonstrated applications and reported as curves and tabular values. All processes are full automated. the model inference was completed within 6-45s after the end of imaging data acquisition.
PAT-CNN: Automatic Segmentation and Quantification of Pericardial Adipose Tissue from T2-Weighted Cardiac Magnetic Resonance Images
Nov 09, 2022Abstract:Background: Increased pericardial adipose tissue (PAT) is associated with many types of cardiovascular disease (CVD). Although cardiac magnetic resonance images (CMRI) are often acquired in patients with CVD, there are currently no tools to automatically identify and quantify PAT from CMRI. The aim of this study was to create a neural network to segment PAT from T2-weighted CMRI and explore the correlations between PAT volumes (PATV) and CVD outcomes and mortality. Methods: We trained and tested a deep learning model, PAT-CNN, to segment PAT on T2-weighted cardiac MR images. Using the segmentations from PAT-CNN, we automatically calculated PATV on images from 391 patients. We analysed correlations between PATV and CVD diagnosis and 1-year mortality post-imaging. Results: PAT-CNN was able to accurately segment PAT with Dice score/ Hausdorff distances of 0.74 +- 0.03/27.1 +- 10.9~mm, similar to the values obtained when comparing the segmentations of two independent human observers ($0.76 +- 0.06/21.2 +- 10.3~mm$). Regression models showed that, independently of sex and body-mass index, PATV is significantly positively correlated with a diagnosis of CVD and with 1-year all cause mortality (p-value < 0.01). Conclusions: PAT-CNN can segment PAT from T2-weighted CMR images automatically and accurately. Increased PATV as measured automatically from CMRI is significantly associated with the presence of CVD and can independently predict 1-year mortality.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge