Wangbin Ding
CineMyoPS: Segmenting Myocardial Pathologies from Cine Cardiac MR
Jul 03, 2025



Abstract:Myocardial infarction (MI) is a leading cause of death worldwide. Late gadolinium enhancement (LGE) and T2-weighted cardiac magnetic resonance (CMR) imaging can respectively identify scarring and edema areas, both of which are essential for MI risk stratification and prognosis assessment. Although combining complementary information from multi-sequence CMR is useful, acquiring these sequences can be time-consuming and prohibitive, e.g., due to the administration of contrast agents. Cine CMR is a rapid and contrast-free imaging technique that can visualize both motion and structural abnormalities of the myocardium induced by acute MI. Therefore, we present a new end-to-end deep neural network, referred to as CineMyoPS, to segment myocardial pathologies, \ie scars and edema, solely from cine CMR images. Specifically, CineMyoPS extracts both motion and anatomy features associated with MI. Given the interdependence between these features, we design a consistency loss (resembling the co-training strategy) to facilitate their joint learning. Furthermore, we propose a time-series aggregation strategy to integrate MI-related features across the cardiac cycle, thereby enhancing segmentation accuracy for myocardial pathologies. Experimental results on a multi-center dataset demonstrate that CineMyoPS achieves promising performance in myocardial pathology segmentation, motion estimation, and anatomy segmentation.
Aligning Multi-Sequence CMR Towards Fully Automated Myocardial Pathology Segmentation
Feb 07, 2023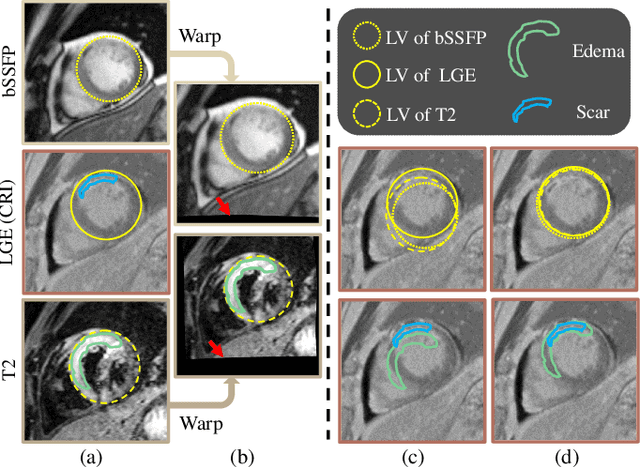
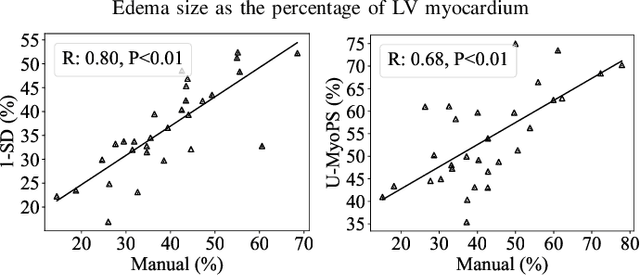
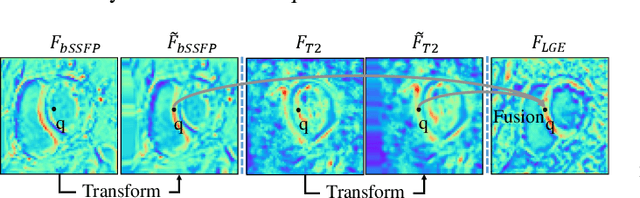
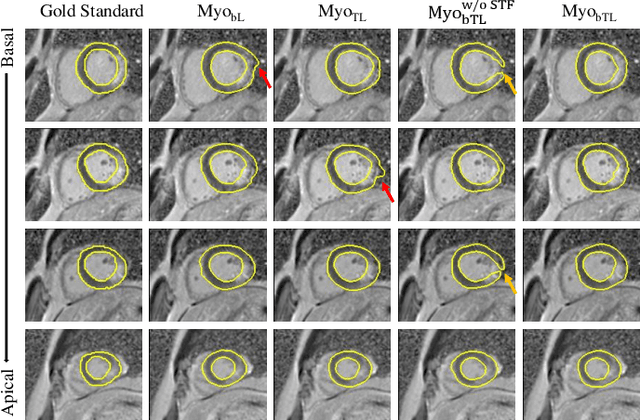
Abstract:Myocardial pathology segmentation (MyoPS) is critical for the risk stratification and treatment planning of myocardial infarction (MI). Multi-sequence cardiac magnetic resonance (MS-CMR) images can provide valuable information. For instance, balanced steady-state free precession cine sequences present clear anatomical boundaries, while late gadolinium enhancement and T2-weighted CMR sequences visualize myocardial scar and edema of MI, respectively. Existing methods usually fuse anatomical and pathological information from different CMR sequences for MyoPS, but assume that these images have been spatially aligned. However, MS-CMR images are usually unaligned due to the respiratory motions in clinical practices, which poses additional challenges for MyoPS. This work presents an automatic MyoPS framework for unaligned MS-CMR images. Specifically, we design a combined computing model for simultaneous image registration and information fusion, which aggregates multi-sequence features into a common space to extract anatomical structures (i.e., myocardium). Consequently, we can highlight the informative regions in the common space via the extracted myocardium to improve MyoPS performance, considering the spatial relationship between myocardial pathologies and myocardium. Experiments on a private MS-CMR dataset and a public dataset from the MYOPS2020 challenge show that our framework could achieve promising performance for fully automatic MyoPS.
Multi-Modality Cardiac Image Computing: A Survey
Aug 26, 2022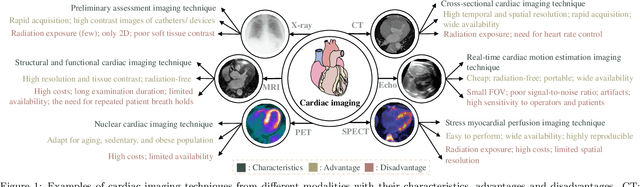
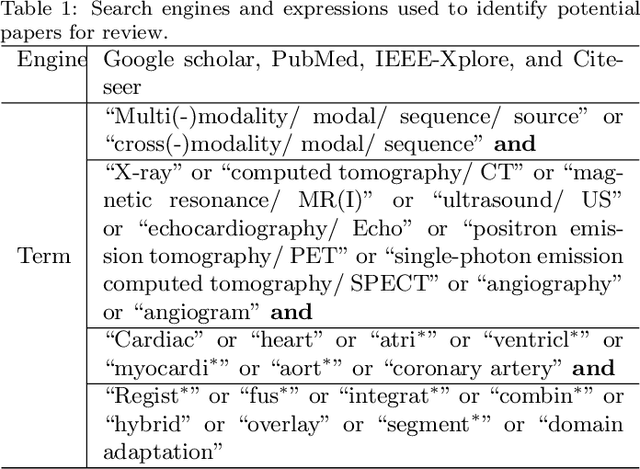
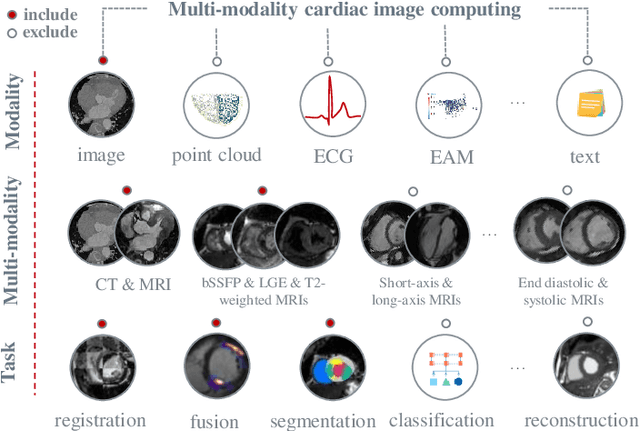
Abstract:Multi-modality cardiac imaging plays a key role in the management of patients with cardiovascular diseases. It allows a combination of complementary anatomical, morphological and functional information, increases diagnosis accuracy, and improves the efficacy of cardiovascular interventions and clinical outcomes. Fully-automated processing and quantitative analysis of multi-modality cardiac images could have a direct impact on clinical research and evidence-based patient management. However, these require overcoming significant challenges including inter-modality misalignment and finding optimal methods to integrate information from different modalities. This paper aims to provide a comprehensive review of multi-modality imaging in cardiology, the computing methods, the validation strategies, the related clinical workflows and future perspectives. For the computing methodologies, we have a favored focus on the three tasks, i.e., registration, fusion and segmentation, which generally involve multi-modality imaging data, \textit{either combining information from different modalities or transferring information across modalities}. The review highlights that multi-modality cardiac imaging data has the potential of wide applicability in the clinic, such as trans-aortic valve implantation guidance, myocardial viability assessment, and catheter ablation therapy and its patient selection. Nevertheless, many challenges remain unsolved, such as missing modality, combination of imaging and non-imaging data, and uniform analysis and representation of different modalities. There is also work to do in defining how the well-developed techniques fit in clinical workflows and how much additional and relevant information they introduce. These problems are likely to continue to be an active field of research and the questions to be answered in the future.
Multi-Depth Boundary-Aware Left Atrial Scar Segmentation Network
Aug 08, 2022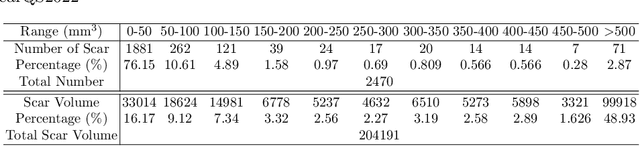
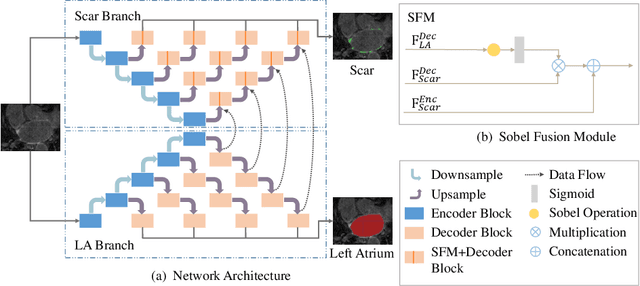
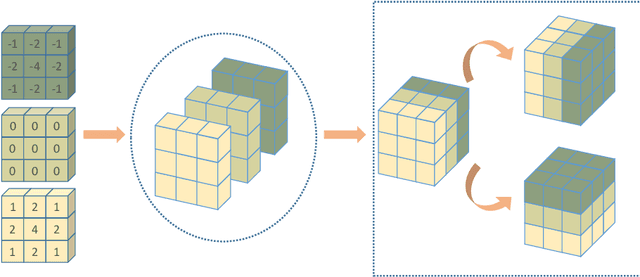

Abstract:Automatic segmentation of left atrial (LA) scars from late gadolinium enhanced CMR images is a crucial step for atrial fibrillation (AF) recurrence analysis. However, delineating LA scars is tedious and error-prone due to the variation of scar shapes. In this work, we propose a boundary-aware LA scar segmentation network, which is composed of two branches to segment LA and LA scars, respectively. We explore the inherent spatial relationship between LA and LA scars. By introducing a Sobel fusion module between the two segmentation branches, the spatial information of LA boundaries can be propagated from the LA branch to the scar branch. Thus, LA scar segmentation can be performed condition on the LA boundaries regions. In our experiments, 40 labeled images were used to train the proposed network, and the remaining 20 labeled images were used for evaluation. The network achieved an average Dice score of 0.608 for LA scar segmentation.
Cross-Modality Multi-Atlas Segmentation Using Deep Neural Networks
Feb 04, 2022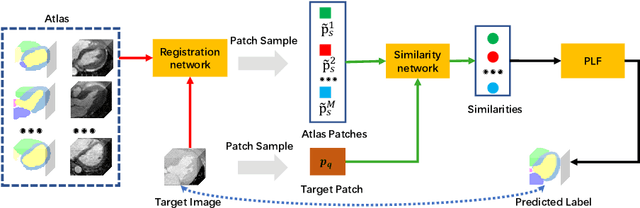
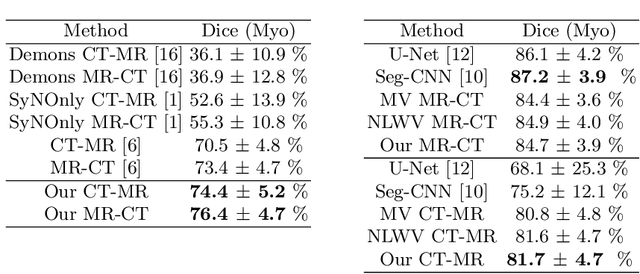
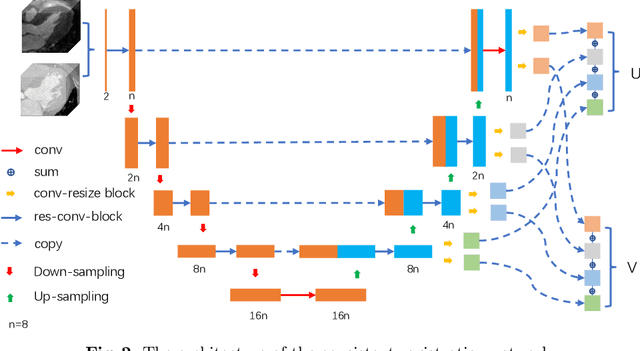
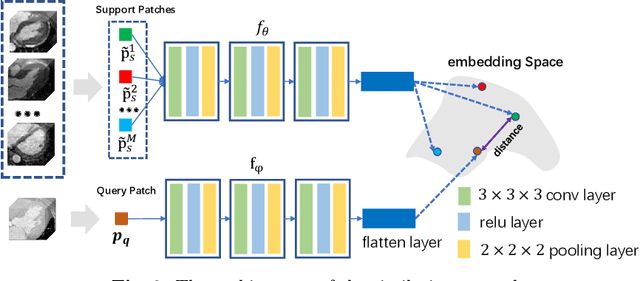
Abstract:Multi-atlas segmentation (MAS) is a promising framework for medical image segmentation. Generally, MAS methods register multiple atlases, i.e., medical images with corresponding labels, to a target image; and the transformed atlas labels can be combined to generate target segmentation via label fusion schemes. Many conventional MAS methods employed the atlases from the same modality as the target image. However, the number of atlases with the same modality may be limited or even missing in many clinical applications. Besides, conventional MAS methods suffer from the computational burden of registration or label fusion procedures. In this work, we design a novel cross-modality MAS framework, which uses available atlases from a certain modality to segment a target image from another modality. To boost the computational efficiency of the framework, both the image registration and label fusion are achieved by well-designed deep neural networks. For the atlas-to-target image registration, we propose a bi-directional registration network (BiRegNet), which can efficiently align images from different modalities. For the label fusion, we design a similarity estimation network (SimNet), which estimates the fusion weight of each atlas by measuring its similarity to the target image. SimNet can learn multi-scale information for similarity estimation to improve the performance of label fusion. The proposed framework was evaluated by the left ventricle and liver segmentation tasks on the MM-WHS and CHAOS datasets, respectively. Results have shown that the framework is effective for cross-modality MAS in both registration and label fusion. The code will be released publicly on \url{https://github.com/NanYoMy/cmmas} once the manuscript is accepted.
Right Ventricular Segmentation from Short- and Long-Axis MRIs via Information Transition
Sep 05, 2021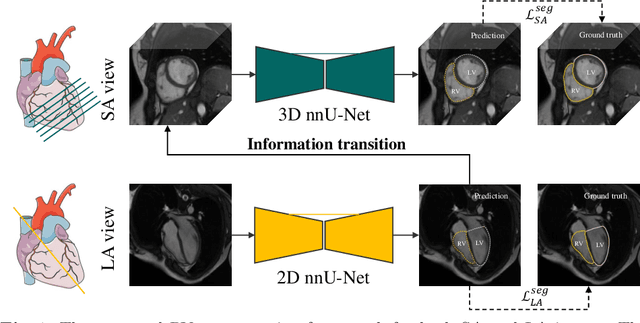

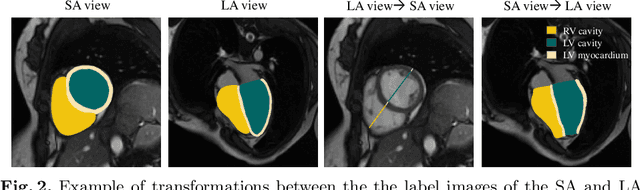
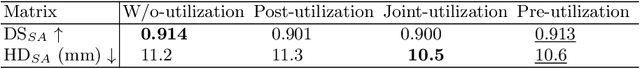
Abstract:Right ventricular (RV) segmentation from magnetic resonance imaging (MRI) is a crucial step for cardiac morphology and function analysis. However, automatic RV segmentation from MRI is still challenging, mainly due to the heterogeneous intensity, the complex variable shapes, and the unclear RV boundary. Moreover, current methods for the RV segmentation tend to suffer from performance degradation at the basal and apical slices of MRI. In this work, we propose an automatic RV segmentation framework, where the information from long-axis (LA) views is utilized to assist the segmentation of short-axis (SA) views via information transition. Specifically, we employed the transformed segmentation from LA views as a prior information, to extract the ROI from SA views for better segmentation. The information transition aims to remove the surrounding ambiguous regions in the SA views. %, such as the tricuspid valve regions. We tested our model on a public dataset with 360 multi-center, multi-vendor and multi-disease subjects that consist of both LA and SA MRIs. Our experimental results show that including LA views can be effective to improve the accuracy of the SA segmentation. Our model is publicly available at https://github.com/NanYoMy/MMs-2.
Unsupervised MMRegNet based on Spatially Encoded Gradient Information
May 16, 2021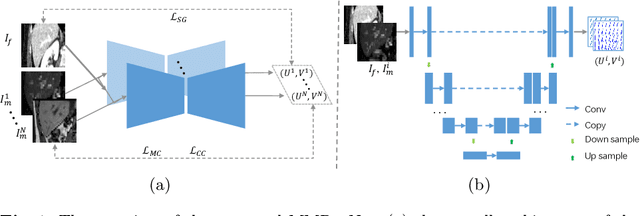
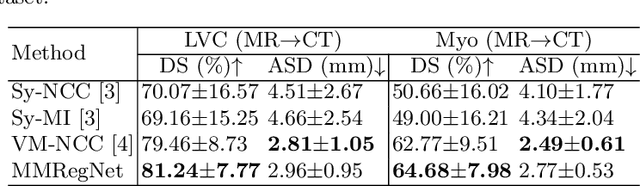
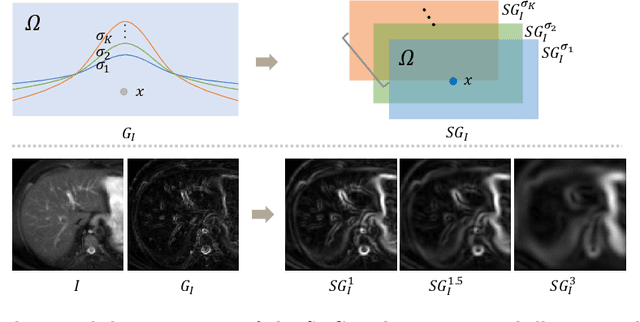
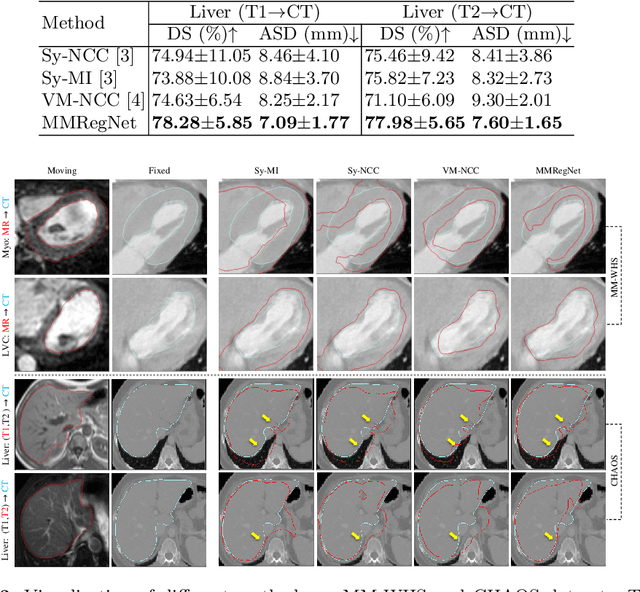
Abstract:Multi-modality medical images can provide relevant and complementary anatomical information for a target (organ, tumor or tissue). Registering the multi-modality images to a common space can fuse these comprehensive information, and bring convenience for clinical application. Recently, neural networks have been widely investigated to boost registration methods. However, it is still challenging to develop a multi-modality registration network due to the lack of robust criteria for network training. Besides, most existing registration networks mainly focus on pairwise registration, and can hardly be applicable for multiple image scenarios. In this work, we propose a multi-modality registration network (MMRegNet), which can jointly register multiple images with different modalities to a target image. Meanwhile, we present spatially encoded gradient information to train the MMRegNet in an unsupervised manner. The proposed network was evaluated on two datasets, i.e, MM-WHS 2017 and CHAOS 2019. The results show that the proposed network can achieve promising performance for cardiac left ventricle and liver registration tasks. Source code is released publicly on github.
Interpretative Computer-aided Lung Cancer Diagnosis: from Radiology Analysis to Malignancy Evaluation
Feb 22, 2021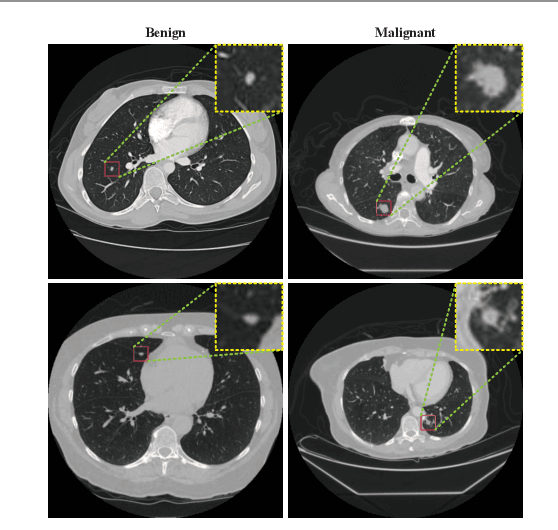
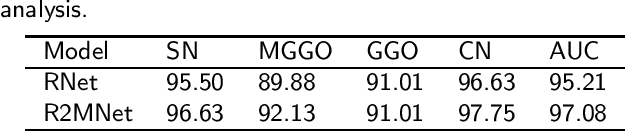
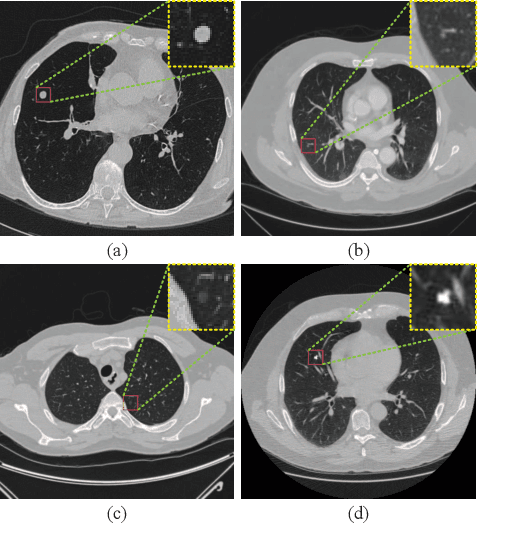
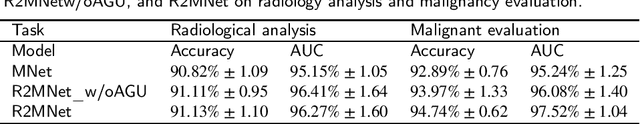
Abstract:Background and Objective:Computer-aided diagnosis (CAD) systems promote diagnosis effectiveness and alleviate pressure of radiologists. A CAD system for lung cancer diagnosis includes nodule candidate detection and nodule malignancy evaluation. Recently, deep learning-based pulmonary nodule detection has reached satisfactory performance ready for clinical application. However, deep learning-based nodule malignancy evaluation depends on heuristic inference from low-dose computed tomography volume to malignant probability, which lacks clinical cognition. Methods:In this paper, we propose a joint radiology analysis and malignancy evaluation network (R2MNet) to evaluate the pulmonary nodule malignancy via radiology characteristics analysis. Radiological features are extracted as channel descriptor to highlight specific regions of the input volume that are critical for nodule malignancy evaluation. In addition, for model explanations, we propose channel-dependent activation mapping to visualize the features and shed light on the decision process of deep neural network. Results:Experimental results on the LIDC-IDRI dataset demonstrate that the proposed method achieved area under curve of 96.27% on nodule radiology analysis and AUC of 97.52% on nodule malignancy evaluation. In addition, explanations of CDAM features proved that the shape and density of nodule regions were two critical factors that influence a nodule to be inferred as malignant, which conforms with the diagnosis cognition of experienced radiologists. Conclusion:Incorporating radiology analysis with nodule malignant evaluation, the network inference process conforms to the diagnostic procedure of radiologists and increases the confidence of evaluation results. Besides, model interpretation with CDAM features shed light on the regions which DNNs focus on when they estimate nodule malignancy probabilities.
Brain Tumor Segmentation Network Using Attention-based Fusion and Spatial Relationship Constraint
Oct 31, 2020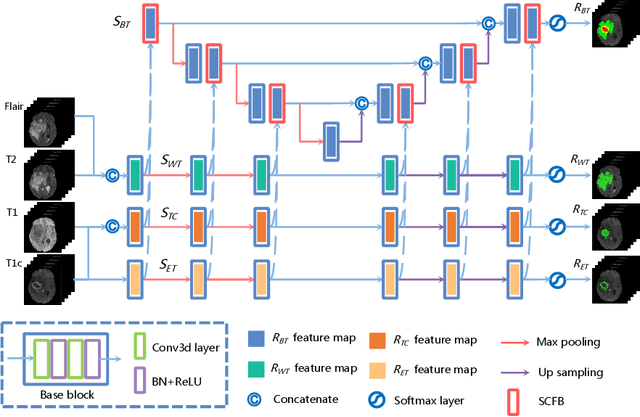
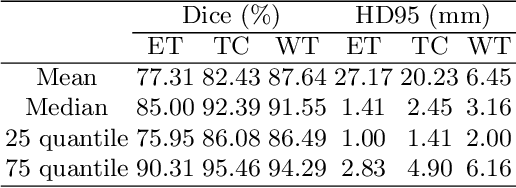
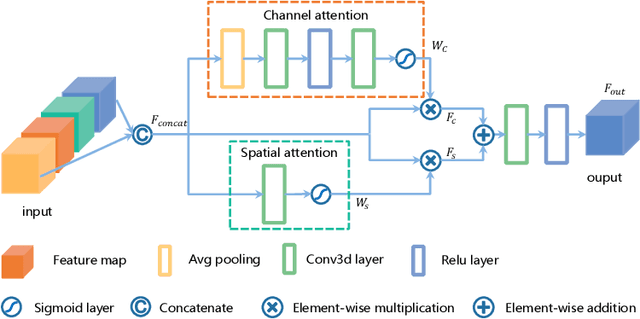
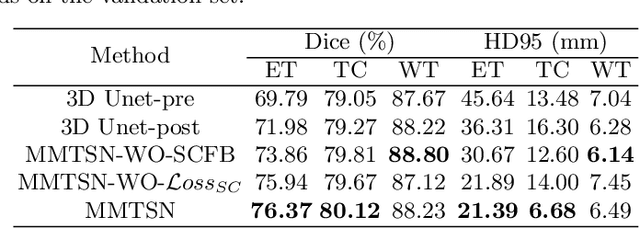
Abstract:Delineating the brain tumor from magnetic resonance (MR) images is critical for the treatment of gliomas. However, automatic delineation is challenging due to the complex appearance and ambiguous outlines of tumors. Considering that multi-modal MR images can reflect different tumor biological properties, we develop a novel multi-modal tumor segmentation network (MMTSN) to robustly segment brain tumors based on multi-modal MR images. The MMTSN is composed of three sub-branches and a main branch. Specifically, the sub-branches are used to capture different tumor features from multi-modal images, while in the main branch, we design a spatial-channel fusion block (SCFB) to effectively aggregate multi-modal features. Additionally, inspired by the fact that the spatial relationship between sub-regions of tumor is relatively fixed, e.g., the enhancing tumor is always in the tumor core, we propose a spatial loss to constrain the relationship between different sub-regions of tumor. We evaluate our method on the test set of multi-modal brain tumor segmentation challenge 2020 (BraTs2020). The method achieves 0.8764, 0.8243 and 0.773 dice score for whole tumor, tumor core and enhancing tumor, respectively.
Random Style Transfer based Domain Generalization Networks Integrating Shape and Spatial Information
Sep 03, 2020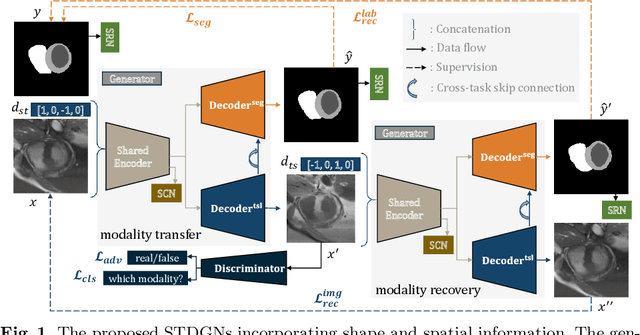
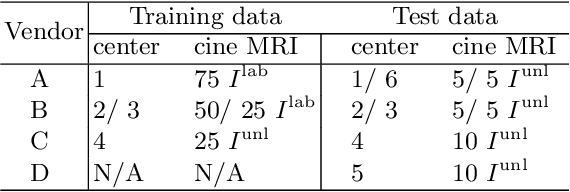
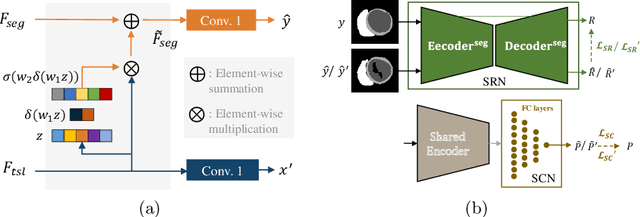
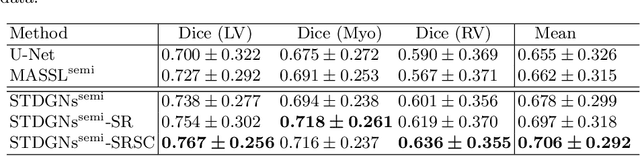
Abstract:Deep learning (DL)-based models have demonstrated good performance in medical image segmentation. However, the models trained on a known dataset often fail when performed on an unseen dataset collected from different centers, vendors and disease populations. In this work, we present a random style transfer network to tackle the domain generalization problem for multi-vendor and center cardiac image segmentation. Style transfer is used to generate training data with a wider distribution/ heterogeneity, namely domain augmentation. As the target domain could be unknown, we randomly generate a modality vector for the target modality in the style transfer stage, to simulate the domain shift for unknown domains. The model can be trained in a semi-supervised manner by simultaneously optimizing a supervised segmentation and an unsupervised style translation objective. Besides, the framework incorporates the spatial information and shape prior of the target by introducing two regularization terms. We evaluated the proposed framework on 40 subjects from the M\&Ms challenge2020, and obtained promising performance in the segmentation for data from unknown vendors and centers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge