Bin Zheng
School of Electrical and Computer Engineering, University of Oklahoma, Norman, OK, USA
Fronthaul-Efficient Distributed Cooperative 3D Positioning with Quantized Latent CSI Embeddings
Jan 31, 2026Abstract:High-precision three-dimensional (3D) positioning in dense urban non-line-of-sight (NLOS) environments benefits significantly from cooperation among multiple distributed base stations (BSs). However, forwarding raw CSI from multiple BSs to a central unit (CU) incurs prohibitive fronthaul overhead, which limits scalable cooperative positioning in practice. This paper proposes a learning-based edge-cloud cooperative positioning framework under limited-capacity fronthaul constraints. In the proposed architecture, a neural network is deployed at each BS to compress the locally estimated CSI into a quantized representation subject to a fixed fronthaul payload. The quantized CSI is transmitted to the CU, which performs cooperative 3D positioning by jointly processing the compressed CSI received from multiple BSs. The proposed framework adopts a two-stage training strategy consisting of self-supervised local training at the BSs and end-to-end joint training for positioning at the CU. Simulation results based on a 3.5~GHz 5G NR compliant urban ray-tracing scenario with six BSs and 20~MHz bandwidth show that the proposed method achieves a mean 3D positioning error of 0.48~m and a 90th-percentile error of 0.83~m, while reducing the fronthaul payload to 6.25% of lossless CSI forwarding. The achieved performance is close to that of cooperative positioning with full CSI exchange.
CMANet: Channel-Masked Attention Network for Cooperative Multi-Base-Station 3D Positioning
Jan 31, 2026Abstract:Achieving ubiquitous high-accuracy localization is crucial for next-generation wireless systems, yet remains challenging in multipath-rich urban environments. By exploiting the fine-grained multipath characteristics embedded in channel state information (CSI), more reliable and precise localization can be achieved. To address this, we present CMANet, a multi-BS cooperative positioning architecture that performs feature-level fusion of raw CSI using the proposed Channel Masked Attention (CMA) mechanism. The CMA encoder injects a physically grounded prior--per-BS channel gain--into the attention weights, thus emphasizing reliable links and suppressing spurious multipath. A lightweight LSTM decoder then treats subcarriers as a sequence to accumulate frequency-domain evidence into a final 3D position estimate. In a typical 5G NR-compliant urban simulation, CMANet achieves less than 0.5m median error and 1.0m 90th-percentile error, outperforming state-of-the-art benchmarks. Ablations verify the necessity of CMA and frequency accumulation. CMANet is edge-deployable and exemplifies an Integrated Sensing and Communication (ISAC)-aligned, cooperative paradigm for multi-BS CSI positioning.
Mammo-CLIP: Leveraging Contrastive Language-Image Pre-training (CLIP) for Enhanced Breast Cancer Diagnosis with Multi-view Mammography
Apr 24, 2024Abstract:Although fusion of information from multiple views of mammograms plays an important role to increase accuracy of breast cancer detection, developing multi-view mammograms-based computer-aided diagnosis (CAD) schemes still faces challenges and no such CAD schemes have been used in clinical practice. To overcome the challenges, we investigate a new approach based on Contrastive Language-Image Pre-training (CLIP), which has sparked interest across various medical imaging tasks. By solving the challenges in (1) effectively adapting the single-view CLIP for multi-view feature fusion and (2) efficiently fine-tuning this parameter-dense model with limited samples and computational resources, we introduce Mammo-CLIP, the first multi-modal framework to process multi-view mammograms and corresponding simple texts. Mammo-CLIP uses an early feature fusion strategy to learn multi-view relationships in four mammograms acquired from the CC and MLO views of the left and right breasts. To enhance learning efficiency, plug-and-play adapters are added into CLIP image and text encoders for fine-tuning parameters and limiting updates to about 1% of the parameters. For framework evaluation, we assembled two datasets retrospectively. The first dataset, comprising 470 malignant and 479 benign cases, was used for few-shot fine-tuning and internal evaluation of the proposed Mammo-CLIP via 5-fold cross-validation. The second dataset, including 60 malignant and 294 benign cases, was used to test generalizability of Mammo-CLIP. Study results show that Mammo-CLIP outperforms the state-of-art cross-view transformer in AUC (0.841 vs. 0.817, 0.837 vs. 0.807) on both datasets. It also surpasses previous two CLIP-based methods by 20.3% and 14.3%. This study highlights the potential of applying the finetuned vision-language models for developing next-generation, image-text-based CAD schemes of breast cancer.
Transformers Improve Breast Cancer Diagnosis from Unregistered Multi-View Mammograms
Jun 21, 2022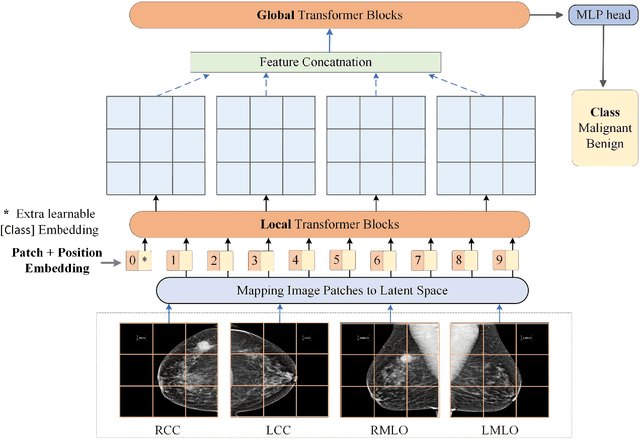
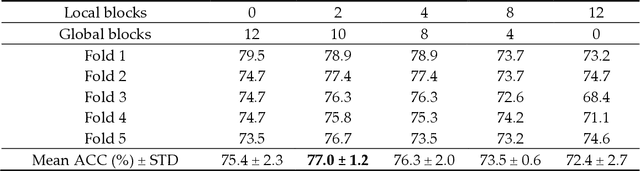
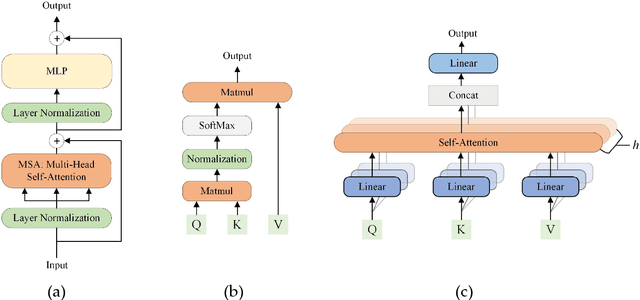
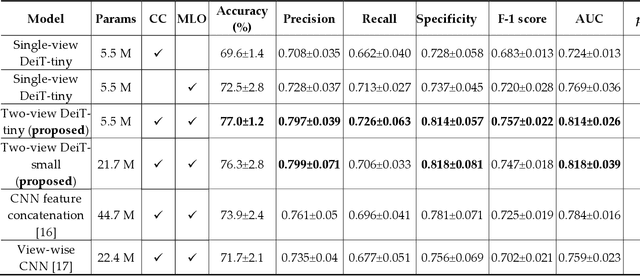
Abstract:Deep convolutional neural networks (CNNs) have been widely used in various medical imaging tasks. However, due to the intrinsic locality of convolution operation, CNNs generally cannot model long-range dependencies well, which are important for accurately identifying or mapping corresponding breast lesion features computed from unregistered multiple mammograms. This motivates us to leverage the architecture of Multi-view Vision Transformers to capture long-range relationships of multiple mammograms from the same patient in one examination. For this purpose, we employ local Transformer blocks to separately learn patch relationships within four mammograms acquired from two-view (CC/MLO) of two-side (right/left) breasts. The outputs from different views and sides are concatenated and fed into global Transformer blocks, to jointly learn patch relationships between four images representing two different views of the left and right breasts. To evaluate the proposed model, we retrospectively assembled a dataset involving 949 sets of mammograms, which include 470 malignant cases and 479 normal or benign cases. We trained and evaluated the model using a five-fold cross-validation method. Without any arduous preprocessing steps (e.g., optimal window cropping, chest wall or pectoral muscle removal, two-view image registration, etc.), our four-image (two-view-two-side) Transformer-based model achieves case classification performance with an area under ROC curve (AUC = 0.818), which significantly outperforms AUC = 0.784 achieved by the state-of-the-art multi-view CNNs (p = 0.009). It also outperforms two one-view-two-side models that achieve AUC of 0.724 (CC view) and 0.769 (MLO view), respectively. The study demonstrates the potential of using Transformers to develop high-performing computer-aided diagnosis schemes that combine four mammograms.
Virtual Adversarial Training for Semi-supervised Breast Mass Classification
Jan 25, 2022Abstract:This study aims to develop a novel computer-aided diagnosis (CAD) scheme for mammographic breast mass classification using semi-supervised learning. Although supervised deep learning has achieved huge success across various medical image analysis tasks, its success relies on large amounts of high-quality annotations, which can be challenging to acquire in practice. To overcome this limitation, we propose employing a semi-supervised method, i.e., virtual adversarial training (VAT), to leverage and learn useful information underlying in unlabeled data for better classification of breast masses. Accordingly, our VAT-based models have two types of losses, namely supervised and virtual adversarial losses. The former loss acts as in supervised classification, while the latter loss aims at enhancing model robustness against virtual adversarial perturbation, thus improving model generalizability. To evaluate the performance of our VAT-based CAD scheme, we retrospectively assembled a total of 1024 breast mass images, with equal number of benign and malignant masses. A large CNN and a small CNN were used in this investigation, and both were trained with and without the adversarial loss. When the labeled ratios were 40% and 80%, VAT-based CNNs delivered the highest classification accuracy of 0.740 and 0.760, respectively. The experimental results suggest that the VAT-based CAD scheme can effectively utilize meaningful knowledge from unlabeled data to better classify mammographic breast mass images.
Recent advances and clinical applications of deep learning in medical image analysis
May 27, 2021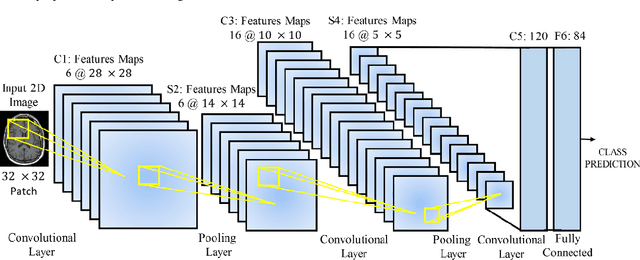
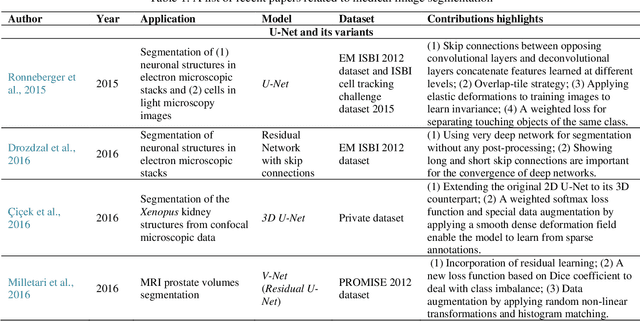
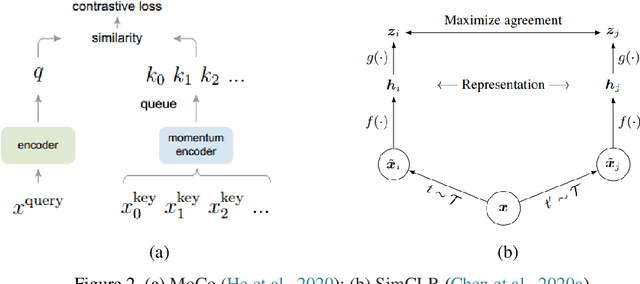
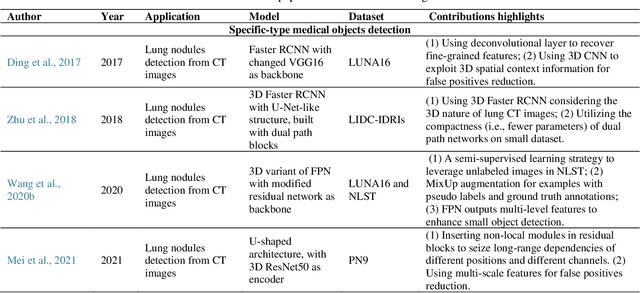
Abstract:Deep learning has become the mainstream technology in computer vision, and it has received extensive research interest in developing new medical image processing algorithms to support disease detection and diagnosis. As compared to conventional machine learning technologies, the major advantage of deep learning is that models can automatically identify and recognize representative features through the hierarchal model architecture, while avoiding the laborious development of hand-crafted features. In this paper, we reviewed and summarized more than 200 recently published papers to provide a comprehensive overview of applying deep learning methods in various medical image analysis tasks. Especially, we emphasize the latest progress and contributions of state-of-the-art unsupervised and semi-supervised deep learning in medical images, which are summarized based on different application scenarios, including lesion classification, segmentation, detection, and image registration. Additionally, we also discussed the major technical challenges and suggested the possible solutions in future research efforts.
Automatic Pulmonary Artery and Vein Separation Algorithm Based on Multitask Classification Network and Topology Reconstruction in Chest CT Images
Mar 22, 2021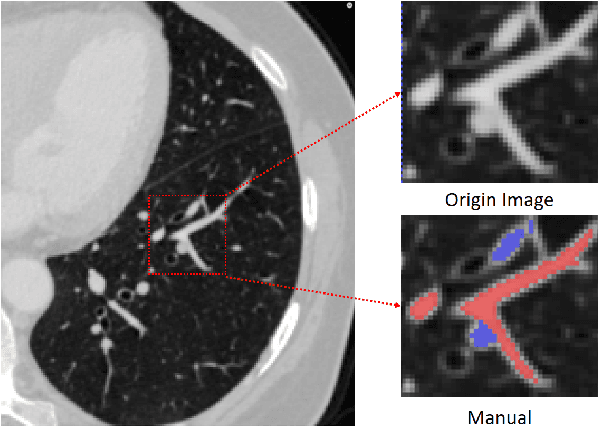
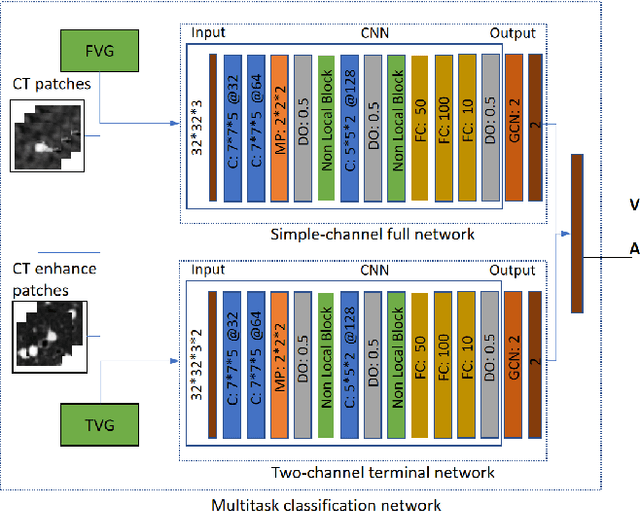
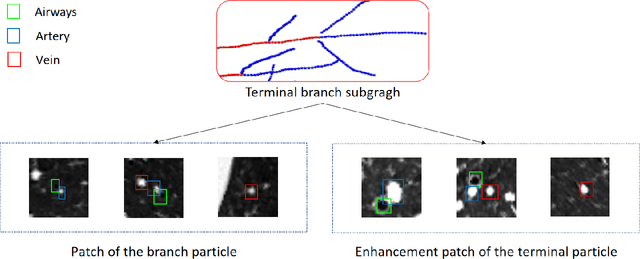
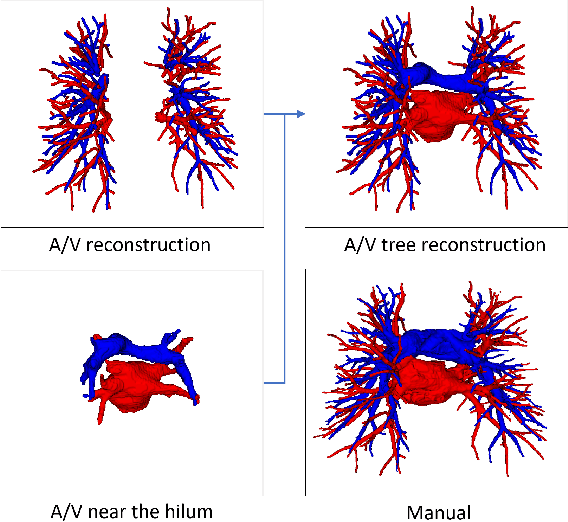
Abstract:With the development of medical computer-aided diagnostic systems, pulmonary artery-vein(A/V) reconstruction plays a crucial role in assisting doctors in preoperative planning for lung cancer surgery. However, distinguishing arterial from venous irrigation in chest CT images remains a challenge due to the similarity and complex structure of the arteries and veins. We propose a novel method for automatic separation of pulmonary arteries and veins from chest CT images. The method consists of three parts. First, global connection information and local feature information are used to construct a complete topological tree and ensure the continuity of vessel reconstruction. Second, the multitask classification network proposed can automatically learn the differences between arteries and veins at different scales to reduce classification errors caused by changes in terminal vessel characteristics. Finally, the topology optimizer considers interbranch and intrabranch topological relationships to maintain spatial consistency to avoid the misclassification of A/V irrigations. We validate the performance of the method on chest CT images. Compared with manual classification, the proposed method achieves an average accuracy of 96.2% on noncontrast chest CT. In addition, the method has been proven to have good generalization, that is, the accuracies of 93.8% and 94.8% are obtained for CT scans from other devices and other modes, respectively. The result of pulmonary artery-vein reconstruction obtained by the proposed method can provide better assistance for preoperative planning of lung cancer surgery.
Coarse-to-fine Airway Segmentation Using Multi information Fusion Network and CNN-based Region Growing
Feb 25, 2021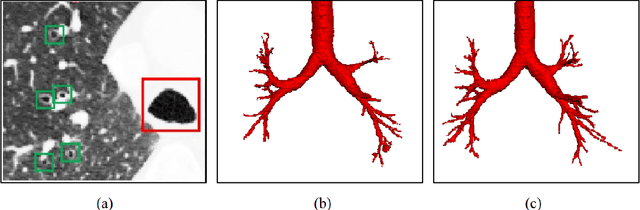
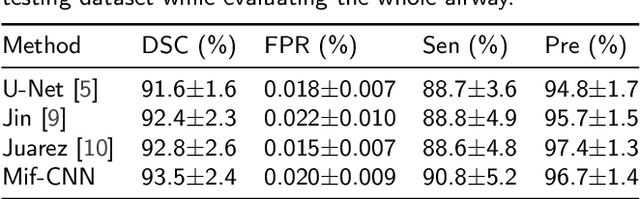
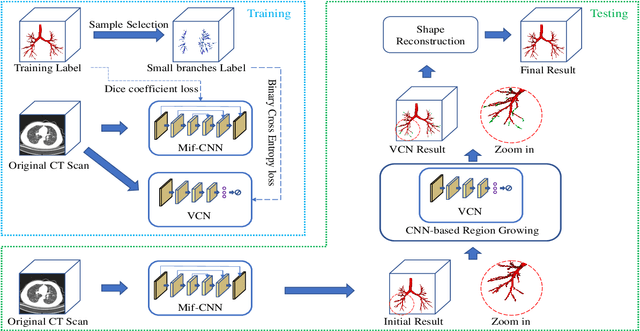
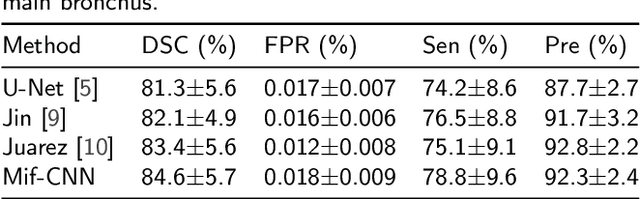
Abstract:Automatic airway segmentation from chest computed tomography (CT) scans plays an important role in pulmonary disease diagnosis and computer-assisted therapy. However, low contrast at peripheral branches and complex tree-like structures remain as two mainly challenges for airway segmentation. Recent research has illustrated that deep learning methods perform well in segmentation tasks. Motivated by these works, a coarse-to-fine segmentation framework is proposed to obtain a complete airway tree. Our framework segments the overall airway and small branches via the multi-information fusion convolution neural network (Mif-CNN) and the CNN-based region growing, respectively. In Mif-CNN, atrous spatial pyramid pooling (ASPP) is integrated into a u-shaped network, and it can expend the receptive field and capture multi-scale information. Meanwhile, boundary and location information are incorporated into semantic information. These information are fused to help Mif-CNN utilize additional context knowledge and useful features. To improve the performance of the segmentation result, the CNN-based region growing method is designed to focus on obtaining small branches. A voxel classification network (VCN), which can entirely capture the rich information around each voxel, is applied to classify the voxels into airway and non-airway. In addition, a shape reconstruction method is used to refine the airway tree.
Interpretative Computer-aided Lung Cancer Diagnosis: from Radiology Analysis to Malignancy Evaluation
Feb 22, 2021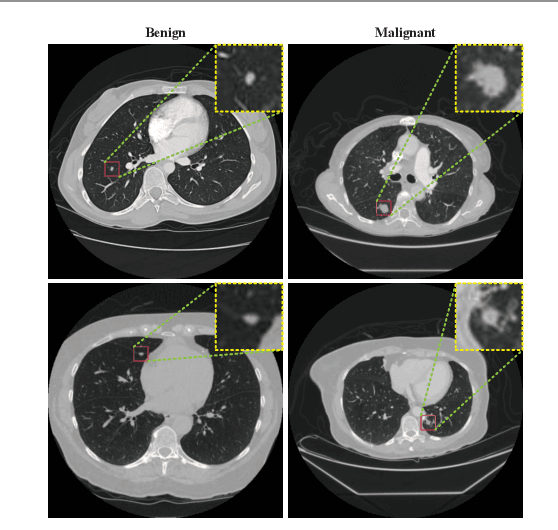
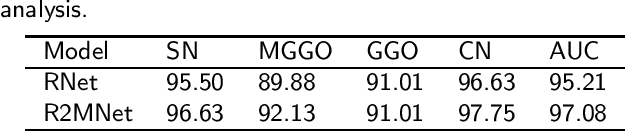
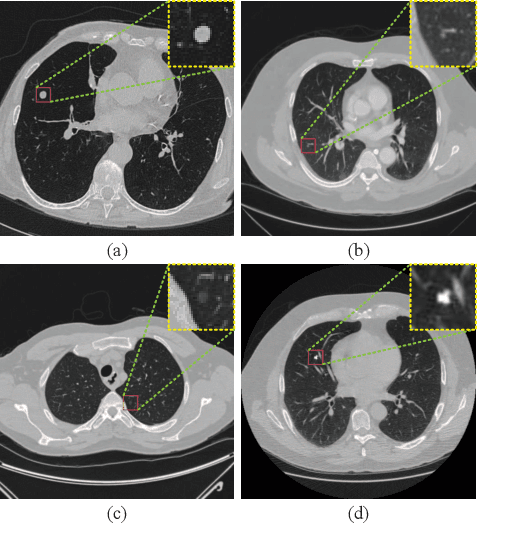
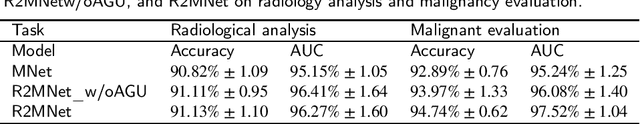
Abstract:Background and Objective:Computer-aided diagnosis (CAD) systems promote diagnosis effectiveness and alleviate pressure of radiologists. A CAD system for lung cancer diagnosis includes nodule candidate detection and nodule malignancy evaluation. Recently, deep learning-based pulmonary nodule detection has reached satisfactory performance ready for clinical application. However, deep learning-based nodule malignancy evaluation depends on heuristic inference from low-dose computed tomography volume to malignant probability, which lacks clinical cognition. Methods:In this paper, we propose a joint radiology analysis and malignancy evaluation network (R2MNet) to evaluate the pulmonary nodule malignancy via radiology characteristics analysis. Radiological features are extracted as channel descriptor to highlight specific regions of the input volume that are critical for nodule malignancy evaluation. In addition, for model explanations, we propose channel-dependent activation mapping to visualize the features and shed light on the decision process of deep neural network. Results:Experimental results on the LIDC-IDRI dataset demonstrate that the proposed method achieved area under curve of 96.27% on nodule radiology analysis and AUC of 97.52% on nodule malignancy evaluation. In addition, explanations of CDAM features proved that the shape and density of nodule regions were two critical factors that influence a nodule to be inferred as malignant, which conforms with the diagnosis cognition of experienced radiologists. Conclusion:Incorporating radiology analysis with nodule malignant evaluation, the network inference process conforms to the diagnostic procedure of radiologists and increases the confidence of evaluation results. Besides, model interpretation with CDAM features shed light on the regions which DNNs focus on when they estimate nodule malignancy probabilities.
Evaluation of company investment value based on machine learning
Sep 30, 2020
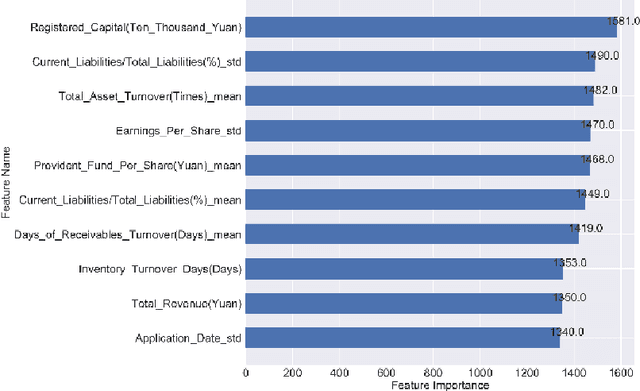
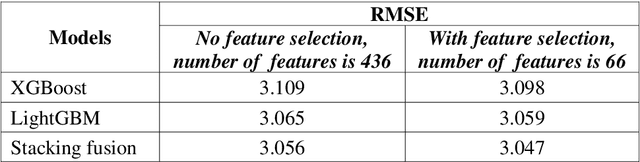
Abstract:In this paper, company investment value evaluation models are established based on comprehensive company information. After data mining and extracting a set of 436 feature parameters, an optimal subset of features is obtained by dimension reduction through tree-based feature selection, followed by the 5-fold cross-validation using XGBoost and LightGBM models. The results show that the Root-Mean-Square Error (RMSE) reached 3.098 and 3.059, respectively. In order to further improve the stability and generalization capability, Bayesian Ridge Regression has been used to train a stacking model based on the XGBoost and LightGBM models. The corresponding RMSE is up to 3.047. Finally, the importance of different features to the LightGBM model is analysed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge