Morteza Heidari
School of Electrical and Computer Engineering, University of Oklahoma, Norman, OK, USA
Deep learning denoising for EOG artifacts removal from EEG signals
Sep 12, 2020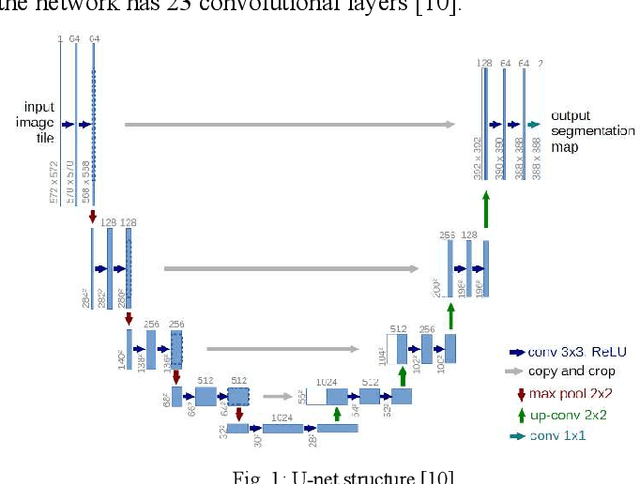
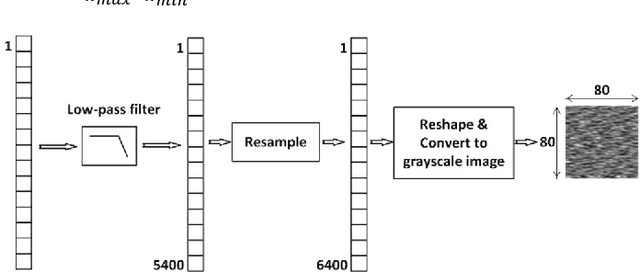

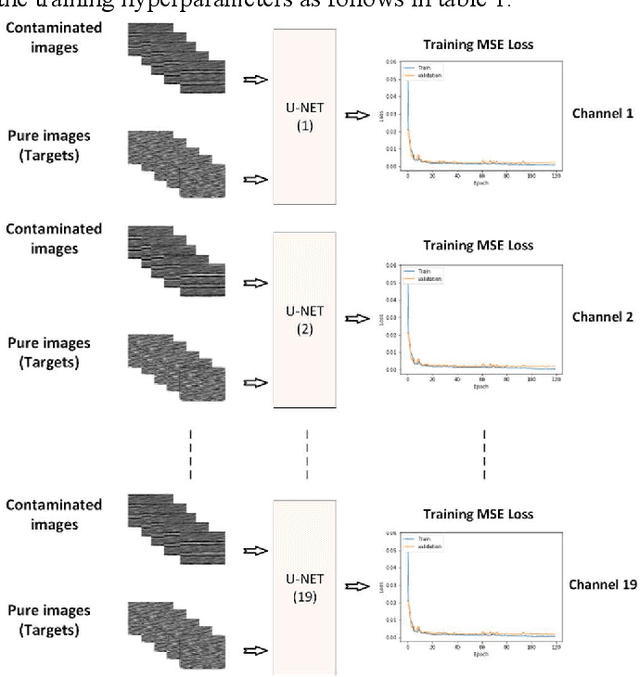
Abstract:There are many sources of interference encountered in the electroencephalogram (EEG) recordings, specifically ocular, muscular, and cardiac artifacts. Rejection of EEG artifacts is an essential process in EEG analysis since such artifacts cause many problems in EEG signals analysis. One of the most challenging issues in EEG denoising processes is removing the ocular artifacts where Electrooculographic (EOG), and EEG signals have an overlap in both frequency and time domains. In this paper, we build and train a deep learning model to deal with this challenge and remove the ocular artifacts effectively. In the proposed scheme, we convert each EEG signal to an image to be fed to a U-NET model, which is a deep learning model usually used in image segmentation tasks. We proposed three different schemes and made our U-NET based models learn to purify contaminated EEG signals similar to the process used in the image segmentation process. The results confirm that one of our schemes can achieve a reliable and promising accuracy to reduce the Mean square error between the target signal (Pure EEGs) and the predicted signal (Purified EEGs).
An approach to human iris recognition using quantitative analysis of image features and machine learning
Sep 12, 2020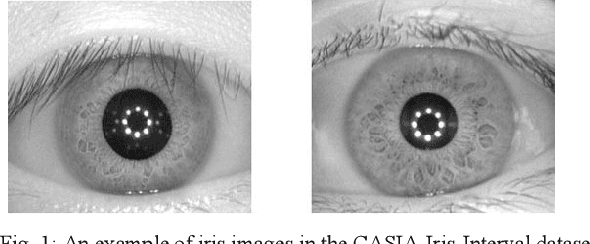
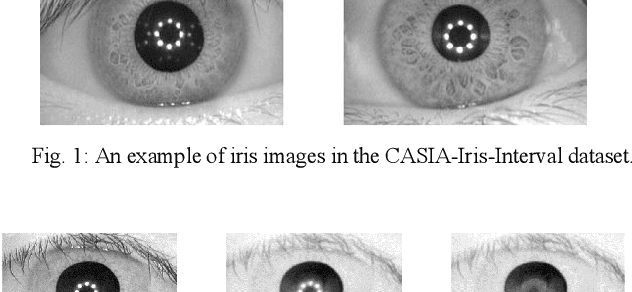


Abstract:The Iris pattern is a unique biological feature for each individual, making it a valuable and powerful tool for human identification. In this paper, an efficient framework for iris recognition is proposed in four steps. (1) Iris segmentation (using a relative total variation combined with Coarse Iris Localization), (2) feature extraction (using Shape&density, FFT, GLCM, GLDM, and Wavelet), (3) feature reduction (employing Kernel-PCA) and (4) classification (applying multi-layer neural network) to classify 2000 iris images of CASIA-Iris-Interval dataset obtained from 200 volunteers. The results confirm that the proposed scheme can provide a reliable prediction with an accuracy of up to 99.64%.
Applying a random projection algorithm to optimize machine learning model for breast lesion classification
Sep 09, 2020



Abstract:Machine learning is widely used in developing computer-aided diagnosis (CAD) schemes of medical images. However, CAD usually computes large number of image features from the targeted regions, which creates a challenge of how to identify a small and optimal feature vector to build robust machine learning models. In this study, we investigate feasibility of applying a random projection algorithm to build an optimal feature vector from the initially CAD-generated large feature pool and improve performance of machine learning model. We assemble a retrospective dataset involving 1,487 cases of mammograms in which 644 cases have confirmed malignant mass lesions and 843 have benign lesions. A CAD scheme is first applied to segment mass regions and initially compute 181 features. Then, support vector machine (SVM) models embedded with several feature dimensionality reduction methods are built to predict likelihood of lesions being malignant. All SVM models are trained and tested using a leave-one-case-out cross-validation method. SVM generates a likelihood score of each segmented mass region depicting on one-view mammogram. By fusion of two scores of the same mass depicting on two-view mammograms, a case-based likelihood score is also evaluated. Comparing with the principle component analyses, nonnegative matrix factorization, and Chi-squared methods, SVM embedded with the random projection algorithm yielded a significantly higher case-based lesion classification performance with the area under ROC curve of 0.84+0.01 (p<0.02). The study demonstrates that the random project algorithm is a promising method to generate optimal feature vectors to help improve performance of machine learning models of medical images.
Applying a random projection algorithm to optimize machine learning model for predicting peritoneal metastasis in gastric cancer patients using CT images
Sep 01, 2020



Abstract:Background and Objective: Non-invasively predicting the risk of cancer metastasis before surgery plays an essential role in determining optimal treatment methods for cancer patients (including who can benefit from neoadjuvant chemotherapy). Although developing radiomics based machine learning (ML) models has attracted broad research interest for this purpose, it often faces a challenge of how to build a highly performed and robust ML model using small and imbalanced image datasets. Methods: In this study, we explore a new approach to build an optimal ML model. A retrospective dataset involving abdominal computed tomography (CT) images acquired from 159 patients diagnosed with gastric cancer is assembled. Among them, 121 cases have peritoneal metastasis (PM), while 38 cases do not have PM. A computer-aided detection (CAD) scheme is first applied to segment primary gastric tumor volumes and initially computes 315 image features. Then, two Gradient Boosting Machine (GBM) models embedded with two different feature dimensionality reduction methods, namely, the principal component analysis (PCA) and a random projection algorithm (RPA) and a synthetic minority oversampling technique, are built to predict the risk of the patients having PM. All GBM models are trained and tested using a leave-one-case-out cross-validation method. Results: Results show that the GBM embedded with RPA yielded a significantly higher prediction accuracy (71.2%) than using PCA (65.2%) (p<0.05). Conclusions: The study demonstrated that CT images of the primary gastric tumors contain discriminatory information to predict the risk of PM, and RPA is a promising method to generate optimal feature vector, improving the performance of ML models of medical images.
Improving performance of CNN to predict likelihood of COVID-19 using chest X-ray images with preprocessing algorithms
Jun 11, 2020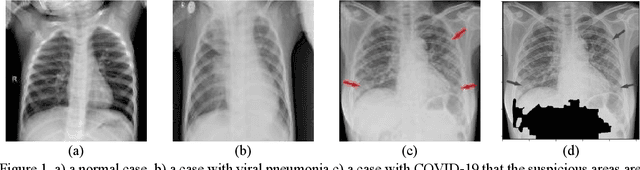
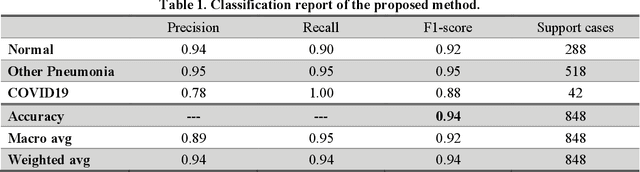
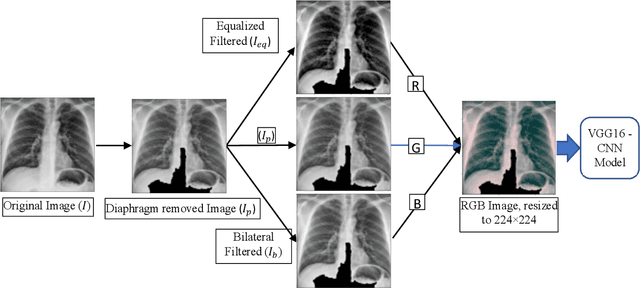
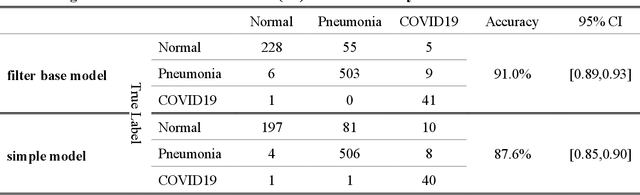
Abstract:As the rapid spread of coronavirus disease (COVID-19) worldwide, chest X-ray radiography has also been used to detect COVID-19 infected pneumonia and assess its severity or monitor its prognosis in the hospitals due to its low cost, low radiation dose, and wide accessibility. However, how to more accurately and efficiently detect COVID-19 infected pneumonia and distinguish it from other community-acquired pneumonia remains a challenge. In order to address this challenge, we in this study develop and test a new computer-aided diagnosis (CAD) scheme. It includes several image pre-processing algorithms to remove diaphragms, normalize image contrast-to-noise ratio, and generate three input images, then links to a transfer learning based convolutional neural network (a VGG16 based CNN model) to classify chest X-ray images into three classes of COVID-19 infected pneumonia, other community-acquired pneumonia and normal (non-pneumonia) cases. To this purpose, a publicly available dataset of 8,474 chest X-ray images is used, which includes 415 confirmed COVID-19 infected pneumonia, 5,179 community-acquired pneumonia, and 2,880 non-pneumonia cases. The dataset is divided into two subsets with 90% and 10% of images in each subset to train and test the CNN-based CAD scheme. The testing results achieve 94.0% of overall accuracy in classifying three classes and 98.6% accuracy in detecting Covid-19 infected cases. Thus, the study demonstrates the feasibility of developing a CAD scheme of chest X-ray images and providing radiologists useful decision-making supporting tools in detecting and diagnosis of COVID-19 infected pneumonia.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge