Mingzhe Hu
Efficient Vision Mamba for MRI Super-Resolution via Hybrid Selective Scanning
Dec 22, 2025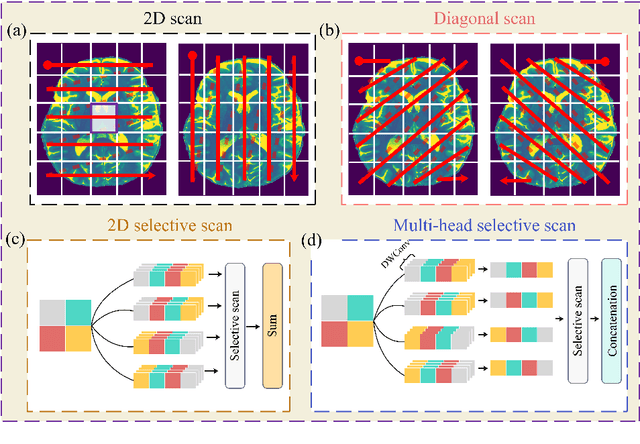
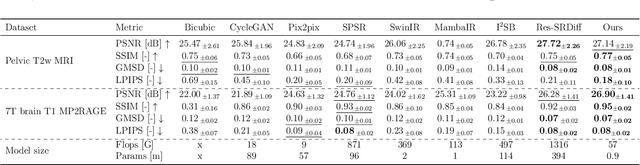
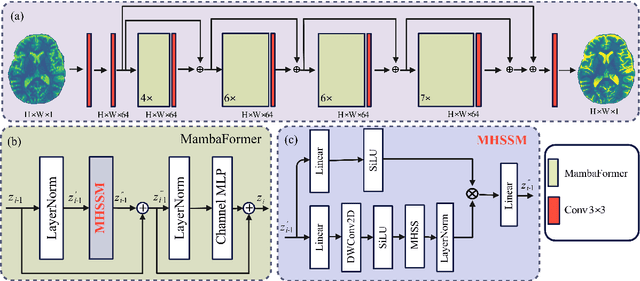

Abstract:Background: High-resolution MRI is critical for diagnosis, but long acquisition times limit clinical use. Super-resolution (SR) can enhance resolution post-scan, yet existing deep learning methods face fidelity-efficiency trade-offs. Purpose: To develop a computationally efficient and accurate deep learning framework for MRI SR that preserves anatomical detail for clinical integration. Materials and Methods: We propose a novel SR framework combining multi-head selective state-space models (MHSSM) with a lightweight channel MLP. The model uses 2D patch extraction with hybrid scanning to capture long-range dependencies. Each MambaFormer block integrates MHSSM, depthwise convolutions, and gated channel mixing. Evaluation used 7T brain T1 MP2RAGE maps (n=142) and 1.5T prostate T2w MRI (n=334). Comparisons included Bicubic interpolation, GANs (CycleGAN, Pix2pix, SPSR), transformers (SwinIR), Mamba (MambaIR), and diffusion models (I2SB, Res-SRDiff). Results: Our model achieved superior performance with exceptional efficiency. For 7T brain data: SSIM=0.951+-0.021, PSNR=26.90+-1.41 dB, LPIPS=0.076+-0.022, GMSD=0.083+-0.017, significantly outperforming all baselines (p<0.001). For prostate data: SSIM=0.770+-0.049, PSNR=27.15+-2.19 dB, LPIPS=0.190+-0.095, GMSD=0.087+-0.013. The framework used only 0.9M parameters and 57 GFLOPs, reducing parameters by 99.8% and computation by 97.5% versus Res-SRDiff, while outperforming SwinIR and MambaIR in accuracy and efficiency. Conclusion: The proposed framework provides an efficient, accurate MRI SR solution, delivering enhanced anatomical detail across datasets. Its low computational demand and state-of-the-art performance show strong potential for clinical translation.
DINOv3 with Test-Time Training for Medical Image Registration
Aug 20, 2025Abstract:Prior medical image registration approaches, particularly learning-based methods, often require large amounts of training data, which constrains clinical adoption. To overcome this limitation, we propose a training-free pipeline that relies on a frozen DINOv3 encoder and test-time optimization of the deformation field in feature space. Across two representative benchmarks, the method is accurate and yields regular deformations. On Abdomen MR-CT, it attained the best mean Dice score (DSC) of 0.790 together with the lowest 95th percentile Hausdorff Distance (HD95) of 4.9+-5.0 and the lowest standard deviation of Log-Jacobian (SDLogJ) of 0.08+-0.02. On ACDC cardiac MRI, it improves mean DSC to 0.769 and reduces SDLogJ to 0.11 and HD95 to 4.8, a marked gain over the initial alignment. The results indicate that operating in a compact foundation feature space at test time offers a practical and general solution for clinical registration without additional training.
Capabilities of GPT-5 on Multimodal Medical Reasoning
Aug 13, 2025Abstract:Recent advances in large language models (LLMs) have enabled general-purpose systems to perform increasingly complex domain-specific reasoning without extensive fine-tuning. In the medical domain, decision-making often requires integrating heterogeneous information sources, including patient narratives, structured data, and medical images. This study positions GPT-5 as a generalist multimodal reasoner for medical decision support and systematically evaluates its zero-shot chain-of-thought reasoning performance on both text-based question answering and visual question answering tasks under a unified protocol. We benchmark GPT-5, GPT-5-mini, GPT-5-nano, and GPT-4o-2024-11-20 against standardized splits of MedQA, MedXpertQA (text and multimodal), MMLU medical subsets, USMLE self-assessment exams, and VQA-RAD. Results show that GPT-5 consistently outperforms all baselines, achieving state-of-the-art accuracy across all QA benchmarks and delivering substantial gains in multimodal reasoning. On MedXpertQA MM, GPT-5 improves reasoning and understanding scores by +29.26% and +26.18% over GPT-4o, respectively, and surpasses pre-licensed human experts by +24.23% in reasoning and +29.40% in understanding. In contrast, GPT-4o remains below human expert performance in most dimensions. A representative case study demonstrates GPT-5's ability to integrate visual and textual cues into a coherent diagnostic reasoning chain, recommending appropriate high-stakes interventions. Our results show that, on these controlled multimodal reasoning benchmarks, GPT-5 moves from human-comparable to above human-expert performance. This improvement may substantially inform the design of future clinical decision-support systems.
Limited-Angle CBCT Reconstruction via Geometry-Integrated Cycle-domain Denoising Diffusion Probabilistic Models
Jun 16, 2025



Abstract:Cone-beam CT (CBCT) is widely used in clinical radiotherapy for image-guided treatment, improving setup accuracy, adaptive planning, and motion management. However, slow gantry rotation limits performance by introducing motion artifacts, blurring, and increased dose. This work aims to develop a clinically feasible method for reconstructing high-quality CBCT volumes from consecutive limited-angle acquisitions, addressing imaging challenges in time- or dose-constrained settings. We propose a limited-angle (LA) geometry-integrated cycle-domain (LA-GICD) framework for CBCT reconstruction, comprising two denoising diffusion probabilistic models (DDPMs) connected via analytic cone-beam forward and back projectors. A Projection-DDPM completes missing projections, followed by back-projection, and an Image-DDPM refines the volume. This dual-domain design leverages complementary priors from projection and image spaces to achieve high-quality reconstructions from limited-angle (<= 90 degrees) scans. Performance was evaluated against full-angle reconstruction. Four board-certified medical physicists conducted assessments. A total of 78 planning CTs in common CBCT geometries were used for training and evaluation. The method achieved a mean absolute error of 35.5 HU, SSIM of 0.84, and PSNR of 29.8 dB, with visibly reduced artifacts and improved soft-tissue clarity. LA-GICD's geometry-aware dual-domain learning, embedded in analytic forward/backward operators, enabled artifact-free, high-contrast reconstructions from a single 90-degree scan, reducing acquisition time and dose four-fold. LA-GICD improves limited-angle CBCT reconstruction with strong data fidelity and anatomical realism. It offers a practical solution for short-arc acquisitions, enhancing CBCT use in radiotherapy by providing clinically applicable images with reduced scan time and dose for more accurate, personalized treatments.
RoMedFormer: A Rotary-Embedding Transformer Foundation Model for 3D Genito-Pelvic Structure Segmentation in MRI and CT
Mar 18, 2025



Abstract:Deep learning-based segmentation of genito-pelvic structures in MRI and CT is crucial for applications such as radiation therapy, surgical planning, and disease diagnosis. However, existing segmentation models often struggle with generalizability across imaging modalities, and anatomical variations. In this work, we propose RoMedFormer, a rotary-embedding transformer-based foundation model designed for 3D female genito-pelvic structure segmentation in both MRI and CT. RoMedFormer leverages self-supervised learning and rotary positional embeddings to enhance spatial feature representation and capture long-range dependencies in 3D medical data. We pre-train our model using a diverse dataset of 3D MRI and CT scans and fine-tune it for downstream segmentation tasks. Experimental results demonstrate that RoMedFormer achieves superior performance segmenting genito-pelvic organs. Our findings highlight the potential of transformer-based architectures in medical image segmentation and pave the way for more transferable segmentation frameworks.
Mammo-CLIP: Leveraging Contrastive Language-Image Pre-training (CLIP) for Enhanced Breast Cancer Diagnosis with Multi-view Mammography
Apr 24, 2024Abstract:Although fusion of information from multiple views of mammograms plays an important role to increase accuracy of breast cancer detection, developing multi-view mammograms-based computer-aided diagnosis (CAD) schemes still faces challenges and no such CAD schemes have been used in clinical practice. To overcome the challenges, we investigate a new approach based on Contrastive Language-Image Pre-training (CLIP), which has sparked interest across various medical imaging tasks. By solving the challenges in (1) effectively adapting the single-view CLIP for multi-view feature fusion and (2) efficiently fine-tuning this parameter-dense model with limited samples and computational resources, we introduce Mammo-CLIP, the first multi-modal framework to process multi-view mammograms and corresponding simple texts. Mammo-CLIP uses an early feature fusion strategy to learn multi-view relationships in four mammograms acquired from the CC and MLO views of the left and right breasts. To enhance learning efficiency, plug-and-play adapters are added into CLIP image and text encoders for fine-tuning parameters and limiting updates to about 1% of the parameters. For framework evaluation, we assembled two datasets retrospectively. The first dataset, comprising 470 malignant and 479 benign cases, was used for few-shot fine-tuning and internal evaluation of the proposed Mammo-CLIP via 5-fold cross-validation. The second dataset, including 60 malignant and 294 benign cases, was used to test generalizability of Mammo-CLIP. Study results show that Mammo-CLIP outperforms the state-of-art cross-view transformer in AUC (0.841 vs. 0.817, 0.837 vs. 0.807) on both datasets. It also surpasses previous two CLIP-based methods by 20.3% and 14.3%. This study highlights the potential of applying the finetuned vision-language models for developing next-generation, image-text-based CAD schemes of breast cancer.
Attention-Driven Lightweight Model for Pigmented Skin Lesion Detection
Aug 04, 2023Abstract:This study presents a lightweight pipeline for skin lesion detection, addressing the challenges posed by imbalanced class distribution and subtle or atypical appearances of some lesions. The pipeline is built around a lightweight model that leverages ghosted features and the DFC attention mechanism to reduce computational complexity while maintaining high performance. The model was trained on the HAM10000 dataset, which includes various types of skin lesions. To address the class imbalance in the dataset, the synthetic minority over-sampling technique and various image augmentation techniques were used. The model also incorporates a knowledge-based loss weighting technique, which assigns different weights to the loss function at the class level and the instance level, helping the model focus on minority classes and challenging samples. This technique involves assigning different weights to the loss function on two levels - the class level and the instance level. By applying appropriate loss weights, the model pays more attention to the minority classes and challenging samples, thus improving its ability to correctly detect and classify different skin lesions. The model achieved an accuracy of 92.4%, a precision of 84.2%, a recall of 86.9%, a f1-score of 85.4% with particularly strong performance in identifying Benign Keratosis-like lesions (BKL) and Nevus (NV). Despite its superior performance, the model's computational cost is considerably lower than some models with less accuracy, making it an optimal solution for real-world applications where both accuracy and efficiency are essential.
BreastSAM: A Study of Segment Anything Model for Breast Tumor Detection in Ultrasound Images
May 21, 2023Abstract:Breast cancer is one of the most common cancers among women worldwide, with early detection significantly increasing survival rates. Ultrasound imaging is a critical diagnostic tool that aids in early detection by providing real-time imaging of the breast tissue. We conducted a thorough investigation of the Segment Anything Model (SAM) for the task of interactive segmentation of breast tumors in ultrasound images. We explored three pre-trained model variants: ViT_h, ViT_l, and ViT_b, among which ViT_l demonstrated superior performance in terms of mean pixel accuracy, Dice score, and IoU score. The significance of prompt interaction in improving the model's segmentation performance was also highlighted, with substantial improvements in performance metrics when prompts were incorporated. The study further evaluated the model's differential performance in segmenting malignant and benign breast tumors, with the model showing exceptional proficiency in both categories, albeit with slightly better performance for benign tumors. Furthermore, we analyzed the impacts of various breast tumor characteristics - size, contrast, aspect ratio, and complexity - on segmentation performance. Our findings reveal that tumor contrast and size positively impact the segmentation result, while complex boundaries pose challenges. The study provides valuable insights for using SAM as a robust and effective algorithm for breast tumor segmentation in ultrasound images.
Polyp-SAM: Transfer SAM for Polyp Segmentation
Apr 29, 2023Abstract:Colon polyps are considered important precursors for colorectal cancer. Automatic segmentation of colon polyps can significantly reduce the misdiagnosis of colon cancer and improve physician annotation efficiency. While many methods have been proposed for polyp segmentation, training large-scale segmentation networks with limited colonoscopy data remains a challenge. Recently, the Segment Anything Model (SAM) has recently gained much attention in both natural and medical image segmentation. SAM demonstrates superior performance in several image benchmarks and therefore shows great potential for medical image segmentation. In this study, we propose Poly-SAM, a finetuned SAM model for polyp segmentation, and compare its performance to several state-of-the-art polyp segmentation models. We also compare two transfer learning strategies of SAM with and without finetuning its encoders. Evaluated on five public datasets, our Polyp-SAM achieves state-of-the-art performance on two datasets and impressive performance on three datasets, with dice scores all above 88%. This study demonstrates the great potential of adapting SAM to medical image segmentation tasks. We plan to release the code and model weights for this paper at: https://github.com/ricklisz/Polyp-SAM.
SkinSAM: Empowering Skin Cancer Segmentation with Segment Anything Model
Apr 27, 2023



Abstract:Skin cancer is a prevalent and potentially fatal disease that requires accurate and efficient diagnosis and treatment. Although manual tracing is the current standard in clinics, automated tools are desired to reduce human labor and improve accuracy. However, developing such tools is challenging due to the highly variable appearance of skin cancers and complex objects in the background. In this paper, we present SkinSAM, a fine-tuned model based on the Segment Anything Model that showed outstanding segmentation performance. The models are validated on HAM10000 dataset which includes 10015 dermatoscopic images. While larger models (ViT_L, ViT_H) performed better than the smaller one (ViT_b), the finetuned model (ViT_b_finetuned) exhibited the greatest improvement, with a Mean pixel accuracy of 0.945, Mean dice score of 0.8879, and Mean IoU score of 0.7843. Among the lesion types, vascular lesions showed the best segmentation results. Our research demonstrates the great potential of adapting SAM to medical image segmentation tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge