Maria Thor
Pretrained hybrid transformer for generalizable cardiac substructures segmentation from contrast and non-contrast CTs in lung and breast cancers
May 16, 2025Abstract:AI automated segmentations for radiation treatment planning (RTP) can deteriorate when applied in clinical cases with different characteristics than training dataset. Hence, we refined a pretrained transformer into a hybrid transformer convolutional network (HTN) to segment cardiac substructures lung and breast cancer patients acquired with varying imaging contrasts and patient scan positions. Cohort I, consisting of 56 contrast-enhanced (CECT) and 124 non-contrast CT (NCCT) scans from patients with non-small cell lung cancers acquired in supine position, was used to create oracle with all 180 training cases and balanced (CECT: 32, NCCT: 32 training) HTN models. Models were evaluated on a held-out validation set of 60 cohort I patients and 66 patients with breast cancer from cohort II acquired in supine (n=45) and prone (n=21) positions. Accuracy was measured using DSC, HD95, and dose metrics. Publicly available TotalSegmentator served as the benchmark. The oracle and balanced models were similarly accurate (DSC Cohort I: 0.80 \pm 0.10 versus 0.81 \pm 0.10; Cohort II: 0.77 \pm 0.13 versus 0.80 \pm 0.12), outperforming TotalSegmentator. The balanced model, using half the training cases as oracle, produced similar dose metrics as manual delineations for all cardiac substructures. This model was robust to CT contrast in 6 out of 8 substructures and patient scan position variations in 5 out of 8 substructures and showed low correlations of accuracy to patient size and age. A HTN demonstrated robustly accurate (geometric and dose metrics) cardiac substructures segmentation from CTs with varying imaging and patient characteristics, one key requirement for clinical use. Moreover, the model combining pretraining with balanced distribution of NCCT and CECT scans was able to provide reliably accurate segmentations under varied conditions with far fewer labeled datasets compared to an oracle model.
RoMedFormer: A Rotary-Embedding Transformer Foundation Model for 3D Genito-Pelvic Structure Segmentation in MRI and CT
Mar 18, 2025



Abstract:Deep learning-based segmentation of genito-pelvic structures in MRI and CT is crucial for applications such as radiation therapy, surgical planning, and disease diagnosis. However, existing segmentation models often struggle with generalizability across imaging modalities, and anatomical variations. In this work, we propose RoMedFormer, a rotary-embedding transformer-based foundation model designed for 3D female genito-pelvic structure segmentation in both MRI and CT. RoMedFormer leverages self-supervised learning and rotary positional embeddings to enhance spatial feature representation and capture long-range dependencies in 3D medical data. We pre-train our model using a diverse dataset of 3D MRI and CT scans and fine-tune it for downstream segmentation tasks. Experimental results demonstrate that RoMedFormer achieves superior performance segmenting genito-pelvic organs. Our findings highlight the potential of transformer-based architectures in medical image segmentation and pave the way for more transferable segmentation frameworks.
Towards Universal Text-driven CT Image Segmentation
Mar 08, 2025Abstract:Computed tomography (CT) is extensively used for accurate visualization and segmentation of organs and lesions. While deep learning models such as convolutional neural networks (CNNs) and vision transformers (ViTs) have significantly improved CT image analysis, their performance often declines when applied to diverse, real-world clinical data. Although foundation models offer a broader and more adaptable solution, their potential is limited due to the challenge of obtaining large-scale, voxel-level annotations for medical images. In response to these challenges, prompting-based models using visual or text prompts have emerged. Visual-prompting methods, such as the Segment Anything Model (SAM), still require significant manual input and can introduce ambiguity when applied to clinical scenarios. Instead, foundation models that use text prompts offer a more versatile and clinically relevant approach. Notably, current text-prompt models, such as the CLIP-Driven Universal Model, are limited to text prompts already encountered during training and struggle to process the complex and diverse scenarios of real-world clinical applications. Instead of fine-tuning models trained from natural imaging, we propose OpenVocabCT, a vision-language model pretrained on large-scale 3D CT images for universal text-driven segmentation. Using the large-scale CT-RATE dataset, we decompose the diagnostic reports into fine-grained, organ-level descriptions using large language models for multi-granular contrastive learning. We evaluate our OpenVocabCT on downstream segmentation tasks across nine public datasets for organ and tumor segmentation, demonstrating the superior performance of our model compared to existing methods. All code, datasets, and models will be publicly released at https://github.com/ricklisz/OpenVocabCT.
Tumor aware recurrent inter-patient deformable image registration of computed tomography scans with lung cancer
Sep 18, 2024Abstract:Background: Voxel-based analysis (VBA) for population level radiotherapy (RT) outcomes modeling requires topology preserving inter-patient deformable image registration (DIR) that preserves tumors on moving images while avoiding unrealistic deformations due to tumors occurring on fixed images. Purpose: We developed a tumor-aware recurrent registration (TRACER) deep learning (DL) method and evaluated its suitability for VBA. Methods: TRACER consists of encoder layers implemented with stacked 3D convolutional long short term memory network (3D-CLSTM) followed by decoder and spatial transform layers to compute dense deformation vector field (DVF). Multiple CLSTM steps are used to compute a progressive sequence of deformations. Input conditioning was applied by including tumor segmentations with 3D image pairs as input channels. Bidirectional tumor rigidity, image similarity, and deformation smoothness losses were used to optimize the network in an unsupervised manner. TRACER and multiple DL methods were trained with 204 3D CT image pairs from patients with lung cancers (LC) and evaluated using (a) Dataset I (N = 308 pairs) with DL segmented LCs, (b) Dataset II (N = 765 pairs) with manually delineated LCs, and (c) Dataset III with 42 LC patients treated with RT. Results: TRACER accurately aligned normal tissues. It best preserved tumors, blackindicated by the smallest tumor volume difference of 0.24\%, 0.40\%, and 0.13 \% and mean square error in CT intensities of 0.005, 0.005, 0.004, computed between original and resampled moving image tumors, for Datasets I, II, and III, respectively. It resulted in the smallest planned RT tumor dose difference computed between original and resampled moving images of 0.01 Gy and 0.013 Gy when using a female and a male reference.
Multiple resolution residual network for automatic thoracic organs-at-risk segmentation from CT
May 31, 2020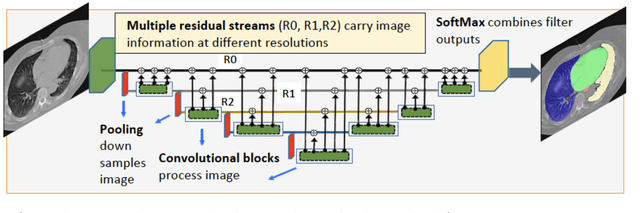
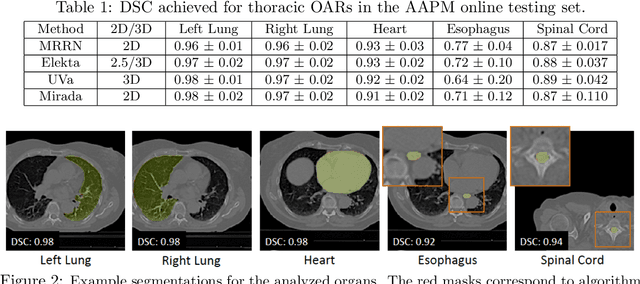
Abstract:We implemented and evaluated a multiple resolution residual network (MRRN) for multiple normal organs-at-risk (OAR) segmentation from computed tomography (CT) images for thoracic radiotherapy treatment (RT) planning. Our approach simultaneously combines feature streams computed at multiple image resolutions and feature levels through residual connections. The feature streams at each level are updated as the images are passed through various feature levels. We trained our approach using 206 thoracic CT scans of lung cancer patients with 35 scans held out for validation to segment the left and right lungs, heart, esophagus, and spinal cord. This approach was tested on 60 CT scans from the open-source AAPM Thoracic Auto-Segmentation Challenge dataset. Performance was measured using the Dice Similarity Coefficient (DSC). Our approach outperformed the best-performing method in the grand challenge for hard-to-segment structures like the esophagus and achieved comparable results for all other structures. Median DSC using our method was 0.97 (interquartile range [IQR]: 0.97-0.98) for the left and right lungs, 0.93 (IQR: 0.93-0.95) for the heart, 0.78 (IQR: 0.76-0.80) for the esophagus, and 0.88 (IQR: 0.86-0.89) for the spinal cord.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge