Yuxiang Lai
AdaFuse: Adaptive Multimodal Fusion for Lung Cancer Risk Prediction via Reinforcement Learning
Jan 30, 2026Abstract:Multimodal fusion has emerged as a promising paradigm for disease diagnosis and prognosis, integrating complementary information from heterogeneous data sources such as medical images, clinical records, and radiology reports. However, existing fusion methods process all available modalities through the network, either treating them equally or learning to assign different contribution weights, leaving a fundamental question unaddressed: for a given patient, should certain modalities be used at all? We present AdaFuse, an adaptive multimodal fusion framework that leverages reinforcement learning (RL) to learn patient-specific modality selection and fusion strategies for lung cancer risk prediction. AdaFuse formulates multimodal fusion as a sequential decision process, where the policy network iteratively decides whether to incorporate an additional modality or proceed to prediction based on the information already acquired. This sequential formulation enables the model to condition each selection on previously observed modalities and terminate early when sufficient information is available, rather than committing to a fixed subset upfront. We evaluate AdaFuse on the National Lung Screening Trial (NLST) dataset. Experimental results demonstrate that AdaFuse achieves the highest AUC (0.762) compared to the best single-modality baseline (0.732), the best fixed fusion strategy (0.759), and adaptive baselines including DynMM (0.754) and MoE (0.742), while using fewer FLOPs than all triple-modality methods. Our work demonstrates the potential of reinforcement learning for personalized multimodal fusion in medical imaging, representing a shift from uniform fusion strategies toward adaptive diagnostic pipelines that learn when to consult additional modalities and when existing information suffices for accurate prediction.
EchoVLM: Measurement-Grounded Multimodal Learning for Echocardiography
Dec 13, 2025Abstract:Echocardiography is the most widely used imaging modality in cardiology, yet its interpretation remains labor-intensive and inherently multimodal, requiring view recognition, quantitative measurements, qualitative assessments, and guideline-based reasoning. While recent vision-language models (VLMs) have achieved broad success in natural images and certain medical domains, their potential in echocardiography has been limited by the lack of large-scale, clinically grounded image-text datasets and the absence of measurement-based reasoning central to echo interpretation. We introduce EchoGround-MIMIC, the first measurement-grounded multimodal echocardiography dataset, comprising 19,065 image-text pairs from 1,572 patients with standardized views, structured measurements, measurement-grounded captions, and guideline-derived disease labels. Building on this resource, we propose EchoVLM, a vision-language model that incorporates two novel pretraining objectives: (i) a view-informed contrastive loss that encodes the view-dependent structure of echocardiographic imaging, and (ii) a negation-aware contrastive loss that distinguishes clinically critical negative from positive findings. Across five types of clinical applications with 36 tasks spanning multimodal disease classification, image-text retrieval, view classification, chamber segmentation, and landmark detection, EchoVLM achieves state-of-the-art performance (86.5% AUC in zero-shot disease classification and 95.1% accuracy in view classification). We demonstrate that clinically grounded multimodal pretraining yields transferable visual representations and establish EchoVLM as a foundation model for end-to-end echocardiography interpretation. We will release EchoGround-MIMIC and the data curation code, enabling reproducibility and further research in multimodal echocardiography interpretation.
MedVista3D: Vision-Language Modeling for Reducing Diagnostic Errors in 3D CT Disease Detection, Understanding and Reporting
Sep 04, 2025Abstract:Radiologic diagnostic errors-under-reading errors, inattentional blindness, and communication failures-remain prevalent in clinical practice. These issues often stem from missed localized abnormalities, limited global context, and variability in report language. These challenges are amplified in 3D imaging, where clinicians must examine hundreds of slices per scan. Addressing them requires systems with precise localized detection, global volume-level reasoning, and semantically consistent natural language reporting. However, existing 3D vision-language models are unable to meet all three needs jointly, lacking local-global understanding for spatial reasoning and struggling with the variability and noise of uncurated radiology reports. We present MedVista3D, a multi-scale semantic-enriched vision-language pretraining framework for 3D CT analysis. To enable joint disease detection and holistic interpretation, MedVista3D performs local and global image-text alignment for fine-grained representation learning within full-volume context. To address report variability, we apply language model rewrites and introduce a Radiology Semantic Matching Bank for semantics-aware alignment. MedVista3D achieves state-of-the-art performance on zero-shot disease classification, report retrieval, and medical visual question answering, while transferring well to organ segmentation and prognosis prediction. Code and datasets will be released.
Patient-Specific Autoregressive Models for Organ Motion Prediction in Radiotherapy
May 17, 2025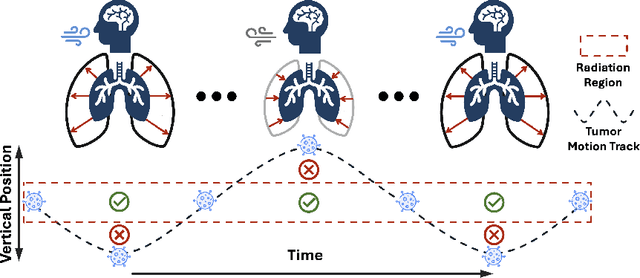
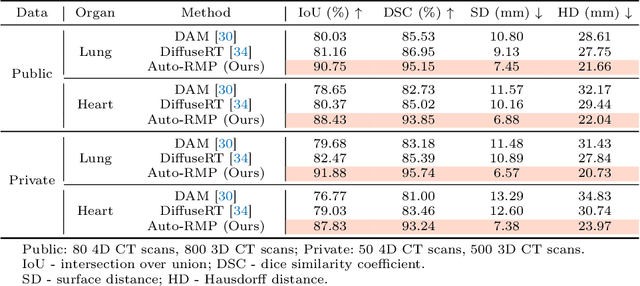
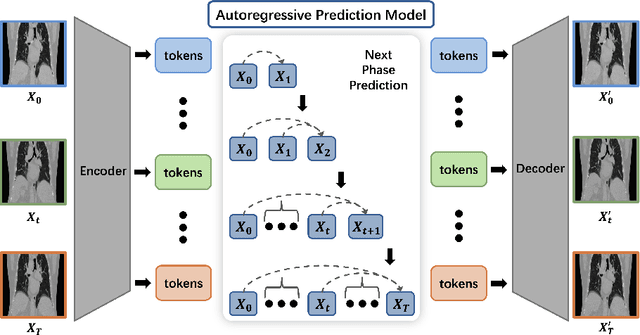
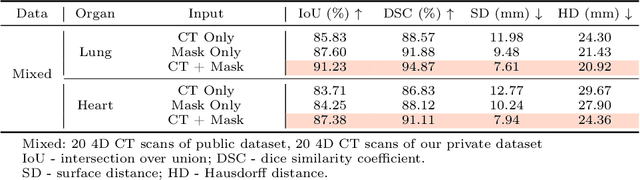
Abstract:Radiotherapy often involves a prolonged treatment period. During this time, patients may experience organ motion due to breathing and other physiological factors. Predicting and modeling this motion before treatment is crucial for ensuring precise radiation delivery. However, existing pre-treatment organ motion prediction methods primarily rely on deformation analysis using principal component analysis (PCA), which is highly dependent on registration quality and struggles to capture periodic temporal dynamics for motion modeling.In this paper, we observe that organ motion prediction closely resembles an autoregressive process, a technique widely used in natural language processing (NLP). Autoregressive models predict the next token based on previous inputs, naturally aligning with our objective of predicting future organ motion phases. Building on this insight, we reformulate organ motion prediction as an autoregressive process to better capture patient-specific motion patterns. Specifically, we acquire 4D CT scans for each patient before treatment, with each sequence comprising multiple 3D CT phases. These phases are fed into the autoregressive model to predict future phases based on prior phase motion patterns. We evaluate our method on a real-world test set of 4D CT scans from 50 patients who underwent radiotherapy at our institution and a public dataset containing 4D CT scans from 20 patients (some with multiple scans), totaling over 1,300 3D CT phases. The performance in predicting the motion of the lung and heart surpasses existing benchmarks, demonstrating its effectiveness in capturing motion dynamics from CT images. These results highlight the potential of our method to improve pre-treatment planning in radiotherapy, enabling more precise and adaptive radiation delivery.
CLS-RL: Image Classification with Rule-Based Reinforcement Learning
Mar 20, 2025



Abstract:Classification is a core task in machine learning. Recent research has shown that although Multimodal Large Language Models (MLLMs) are initially poor at image classification, fine-tuning them with an adequate amount of data can significantly enhance their performance, making them comparable to SOTA classification models. However, acquiring large-scale labeled data is expensive. In this paper, we explore few-shot MLLM classification fine-tuning. We found that SFT can cause severe overfitting issues and may even degrade performance over the zero-shot approach. To address this challenge, inspired by the recent successes in rule-based reinforcement learning, we propose CLS-RL, which uses verifiable signals as reward to fine-tune MLLMs. We discovered that CLS-RL outperforms SFT in most datasets and has a much higher average accuracy on both base-to-new and few-shot learning setting. Moreover, we observed a free-lunch phenomenon for CLS-RL; when models are fine-tuned on a particular dataset, their performance on other distinct datasets may also improve over zero-shot models, even if those datasets differ in distribution and class names. This suggests that RL-based methods effectively teach models the fundamentals of classification. Lastly, inspired by recent works in inference time thinking, we re-examine the `thinking process' during fine-tuning, a critical aspect of RL-based methods, in the context of visual classification. We question whether such tasks require extensive thinking process during fine-tuning, proposing that this may actually detract from performance. Based on this premise, we introduce the No-Thinking-CLS-RL method, which minimizes thinking processes during training by setting an equality accuracy reward. Our findings indicate that, with much less fine-tuning time, No-Thinking-CLS-RL method achieves superior in-domain performance and generalization capabilities than CLS-RL.
Med-R1: Reinforcement Learning for Generalizable Medical Reasoning in Vision-Language Models
Mar 19, 2025



Abstract:Vision-language models (VLMs) have advanced reasoning in natural scenes, but their role in medical imaging remains underexplored. Medical reasoning tasks demand robust image analysis and well-justified answers, posing challenges due to the complexity of medical images. Transparency and trustworthiness are essential for clinical adoption and regulatory compliance. We introduce Med-R1, a framework exploring reinforcement learning (RL) to enhance VLMs' generalizability and trustworthiness in medical reasoning. Leveraging the DeepSeek strategy, we employ Group Relative Policy Optimization (GRPO) to guide reasoning paths via reward signals. Unlike supervised fine-tuning (SFT), which often overfits and lacks generalization, RL fosters robust and diverse reasoning. Med-R1 is evaluated across eight medical imaging modalities: CT, MRI, Ultrasound, Dermoscopy, Fundus Photography, Optical Coherence Tomography (OCT), Microscopy, and X-ray Imaging. Compared to its base model, Qwen2-VL-2B, Med-R1 achieves a 29.94% accuracy improvement and outperforms Qwen2-VL-72B, which has 36 times more parameters. Testing across five question types-modality recognition, anatomy identification, disease diagnosis, lesion grading, and biological attribute analysis Med-R1 demonstrates superior generalization, exceeding Qwen2-VL-2B by 32.06% and surpassing Qwen2-VL-72B in question-type generalization. These findings show that RL improves medical reasoning and enables parameter-efficient models to outperform significantly larger ones. With interpretable reasoning outputs, Med-R1 represents a promising step toward generalizable, trustworthy, and clinically viable medical VLMs.
Towards Universal Text-driven CT Image Segmentation
Mar 08, 2025Abstract:Computed tomography (CT) is extensively used for accurate visualization and segmentation of organs and lesions. While deep learning models such as convolutional neural networks (CNNs) and vision transformers (ViTs) have significantly improved CT image analysis, their performance often declines when applied to diverse, real-world clinical data. Although foundation models offer a broader and more adaptable solution, their potential is limited due to the challenge of obtaining large-scale, voxel-level annotations for medical images. In response to these challenges, prompting-based models using visual or text prompts have emerged. Visual-prompting methods, such as the Segment Anything Model (SAM), still require significant manual input and can introduce ambiguity when applied to clinical scenarios. Instead, foundation models that use text prompts offer a more versatile and clinically relevant approach. Notably, current text-prompt models, such as the CLIP-Driven Universal Model, are limited to text prompts already encountered during training and struggle to process the complex and diverse scenarios of real-world clinical applications. Instead of fine-tuning models trained from natural imaging, we propose OpenVocabCT, a vision-language model pretrained on large-scale 3D CT images for universal text-driven segmentation. Using the large-scale CT-RATE dataset, we decompose the diagnostic reports into fine-grained, organ-level descriptions using large language models for multi-granular contrastive learning. We evaluate our OpenVocabCT on downstream segmentation tasks across nine public datasets for organ and tumor segmentation, demonstrating the superior performance of our model compared to existing methods. All code, datasets, and models will be publicly released at https://github.com/ricklisz/OpenVocabCT.
EEE-Bench: A Comprehensive Multimodal Electrical And Electronics Engineering Benchmark
Nov 03, 2024Abstract:Recent studies on large language models (LLMs) and large multimodal models (LMMs) have demonstrated promising skills in various domains including science and mathematics. However, their capability in more challenging and real-world related scenarios like engineering has not been systematically studied. To bridge this gap, we propose EEE-Bench, a multimodal benchmark aimed at assessing LMMs' capabilities in solving practical engineering tasks, using electrical and electronics engineering (EEE) as the testbed. Our benchmark consists of 2860 carefully curated problems spanning 10 essential subdomains such as analog circuits, control systems, etc. Compared to benchmarks in other domains, engineering problems are intrinsically 1) more visually complex and versatile and 2) less deterministic in solutions. Successful solutions to these problems often demand more-than-usual rigorous integration of visual and textual information as models need to understand intricate images like abstract circuits and system diagrams while taking professional instructions, making them excellent candidates for LMM evaluations. Alongside EEE-Bench, we provide extensive quantitative evaluations and fine-grained analysis of 17 widely-used open and closed-sourced LLMs and LMMs. Our results demonstrate notable deficiencies of current foundation models in EEE, with an average performance ranging from 19.48% to 46.78%. Finally, we reveal and explore a critical shortcoming in LMMs which we term laziness: the tendency to take shortcuts by relying on the text while overlooking the visual context when reasoning for technical image problems. In summary, we believe EEE-Bench not only reveals some noteworthy limitations of LMMs but also provides a valuable resource for advancing research on their application in practical engineering tasks, driving future improvements in their capability to handle complex, real-world scenarios.
Analyzing Tumors by Synthesis
Sep 09, 2024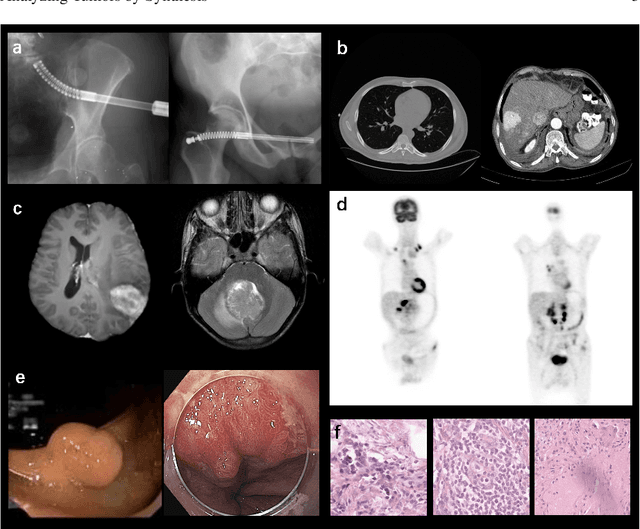
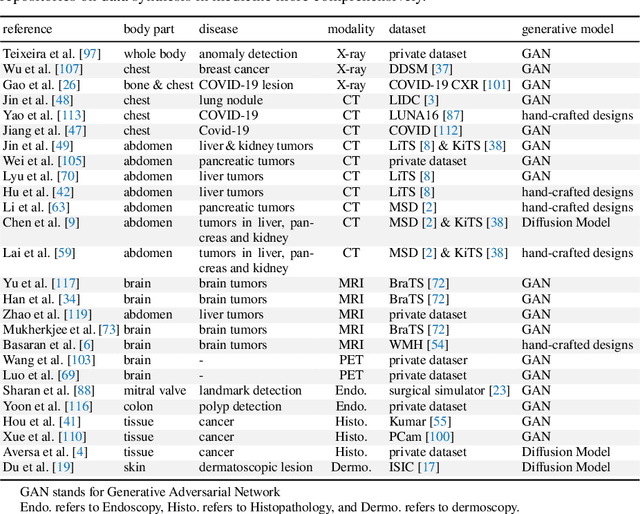
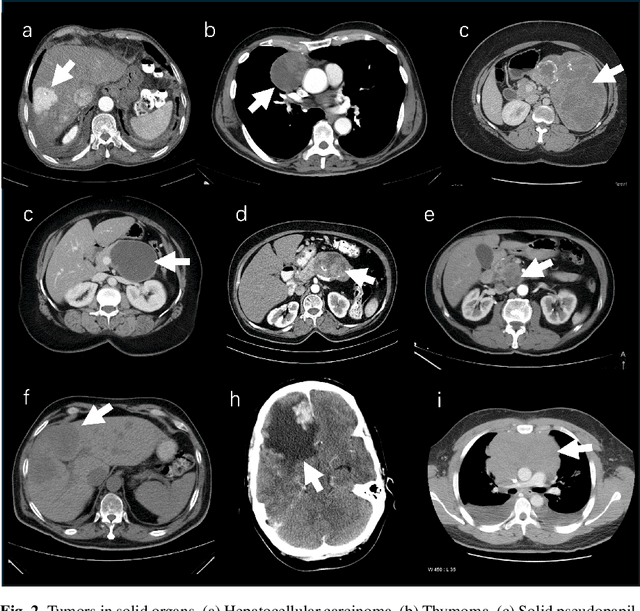
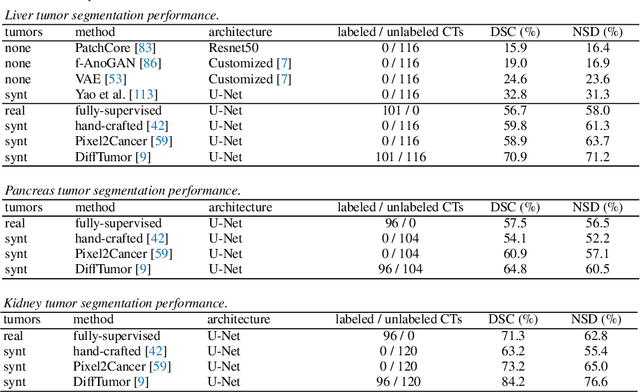
Abstract:Computer-aided tumor detection has shown great potential in enhancing the interpretation of over 80 million CT scans performed annually in the United States. However, challenges arise due to the rarity of CT scans with tumors, especially early-stage tumors. Developing AI with real tumor data faces issues of scarcity, annotation difficulty, and low prevalence. Tumor synthesis addresses these challenges by generating numerous tumor examples in medical images, aiding AI training for tumor detection and segmentation. Successful synthesis requires realistic and generalizable synthetic tumors across various organs. This chapter reviews AI development on real and synthetic data and summarizes two key trends in synthetic data for cancer imaging research: modeling-based and learning-based approaches. Modeling-based methods, like Pixel2Cancer, simulate tumor development over time using generic rules, while learning-based methods, like DiffTumor, learn from a few annotated examples in one organ to generate synthetic tumors in others. Reader studies with expert radiologists show that synthetic tumors can be convincingly realistic. We also present case studies in the liver, pancreas, and kidneys reveal that AI trained on synthetic tumors can achieve performance comparable to, or better than, AI only trained on real data. Tumor synthesis holds significant promise for expanding datasets, enhancing AI reliability, improving tumor detection performance, and preserving patient privacy.
AbdomenAtlas: A Large-Scale, Detailed-Annotated, & Multi-Center Dataset for Efficient Transfer Learning and Open Algorithmic Benchmarking
Jul 23, 2024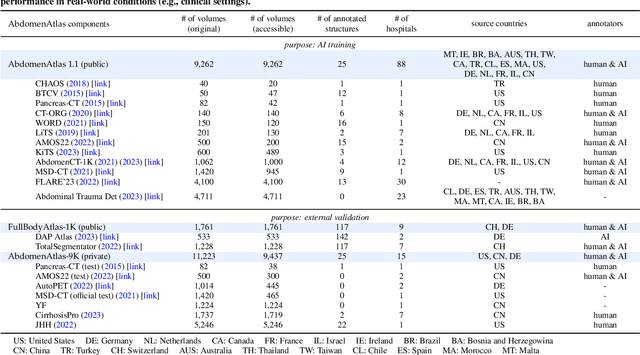


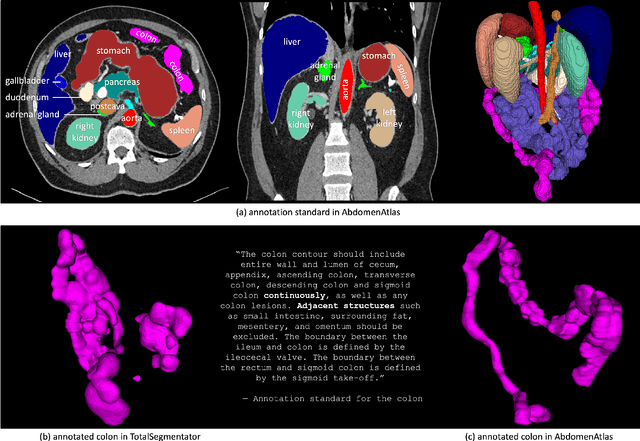
Abstract:We introduce the largest abdominal CT dataset (termed AbdomenAtlas) of 20,460 three-dimensional CT volumes sourced from 112 hospitals across diverse populations, geographies, and facilities. AbdomenAtlas provides 673K high-quality masks of anatomical structures in the abdominal region annotated by a team of 10 radiologists with the help of AI algorithms. We start by having expert radiologists manually annotate 22 anatomical structures in 5,246 CT volumes. Following this, a semi-automatic annotation procedure is performed for the remaining CT volumes, where radiologists revise the annotations predicted by AI, and in turn, AI improves its predictions by learning from revised annotations. Such a large-scale, detailed-annotated, and multi-center dataset is needed for two reasons. Firstly, AbdomenAtlas provides important resources for AI development at scale, branded as large pre-trained models, which can alleviate the annotation workload of expert radiologists to transfer to broader clinical applications. Secondly, AbdomenAtlas establishes a large-scale benchmark for evaluating AI algorithms -- the more data we use to test the algorithms, the better we can guarantee reliable performance in complex clinical scenarios. An ISBI & MICCAI challenge named BodyMaps: Towards 3D Atlas of Human Body was launched using a subset of our AbdomenAtlas, aiming to stimulate AI innovation and to benchmark segmentation accuracy, inference efficiency, and domain generalizability. We hope our AbdomenAtlas can set the stage for larger-scale clinical trials and offer exceptional opportunities to practitioners in the medical imaging community. Codes, models, and datasets are available at https://www.zongweiz.com/dataset
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge