Tao Zhou
Bidirectional Channel-selective Semantic Interaction for Semi-Supervised Medical Segmentation
Jan 09, 2026Abstract:Semi-supervised medical image segmentation is an effective method for addressing scenarios with limited labeled data. Existing methods mainly rely on frameworks such as mean teacher and dual-stream consistency learning. These approaches often face issues like error accumulation and model structural complexity, while also neglecting the interaction between labeled and unlabeled data streams. To overcome these challenges, we propose a Bidirectional Channel-selective Semantic Interaction~(BCSI) framework for semi-supervised medical image segmentation. First, we propose a Semantic-Spatial Perturbation~(SSP) mechanism, which disturbs the data using two strong augmentation operations and leverages unsupervised learning with pseudo-labels from weak augmentations. Additionally, we employ consistency on the predictions from the two strong augmentations to further improve model stability and robustness. Second, to reduce noise during the interaction between labeled and unlabeled data, we propose a Channel-selective Router~(CR) component, which dynamically selects the most relevant channels for information exchange. This mechanism ensures that only highly relevant features are activated, minimizing unnecessary interference. Finally, the Bidirectional Channel-wise Interaction~(BCI) strategy is employed to supplement additional semantic information and enhance the representation of important channels. Experimental results on multiple benchmarking 3D medical datasets demonstrate that the proposed method outperforms existing semi-supervised approaches.
A Novel Deep Learning-Based Coarse-to-Fine Frame Synchronization Method for OTFS Systems
Jan 09, 2026Abstract:Orthogonal time frequency space (OTFS) modulation is a robust candidate waveform for future wireless systems, particularly in high-mobility scenarios, as it effectively mitigates the impact of rapidly time-varying channels by mapping symbols in the delay-Doppler (DD) domain. However, accurate frame synchronization in OTFS systems remains a challenge due to the performance limitations of conventional algorithms. To address this, we propose a low-complexity synchronization method based on a coarse-to-fine deep residual network (ResNet) architecture. Unlike traditional approaches relying on high-overhead preamble structures, our method exploits the intrinsic periodic features of OTFS pilots in the delay-time (DT) domain to formulate synchronization as a hierarchical classification problem. Specifically, the proposed architecture employs a two-stage strategy to first narrow the search space and then pinpoint the precise symbol timing offset (STO), thereby significantly reducing computational complexity while maintaining high estimation accuracy. We construct a comprehensive simulation dataset incorporating diverse channel models and randomized STO to validate the method. Extensive simulation results demonstrate that the proposed method achieves robust signal start detection and superior accuracy compared to conventional benchmarks, particularly in low signal-to-noise ratio (SNR) regimes and high-mobility scenarios.
Tyee: A Unified, Modular, and Fully-Integrated Configurable Toolkit for Intelligent Physiological Health Care
Dec 27, 2025Abstract:Deep learning has shown great promise in physiological signal analysis, yet its progress is hindered by heterogeneous data formats, inconsistent preprocessing strategies, fragmented model pipelines, and non-reproducible experimental setups. To address these limitations, we present Tyee, a unified, modular, and fully-integrated configurable toolkit designed for intelligent physiological healthcare. Tyee introduces three key innovations: (1) a unified data interface and configurable preprocessing pipeline for 12 kinds of signal modalities; (2) a modular and extensible architecture enabling flexible integration and rapid prototyping across tasks; and (3) end-to-end workflow configuration, promoting reproducible and scalable experimentation. Tyee demonstrates consistent practical effectiveness and generalizability, outperforming or matching baselines across all evaluated tasks (with state-of-the-art results on 12 of 13 datasets). The Tyee toolkit is released at https://github.com/SmileHnu/Tyee and actively maintained.
Contrastive Graph Modeling for Cross-Domain Few-Shot Medical Image Segmentation
Dec 25, 2025Abstract:Cross-domain few-shot medical image segmentation (CD-FSMIS) offers a promising and data-efficient solution for medical applications where annotations are severely scarce and multimodal analysis is required. However, existing methods typically filter out domain-specific information to improve generalization, which inadvertently limits cross-domain performance and degrades source-domain accuracy. To address this, we present Contrastive Graph Modeling (C-Graph), a framework that leverages the structural consistency of medical images as a reliable domain-transferable prior. We represent image features as graphs, with pixels as nodes and semantic affinities as edges. A Structural Prior Graph (SPG) layer is proposed to capture and transfer target-category node dependencies and enable global structure modeling through explicit node interactions. Building upon SPG layers, we introduce a Subgraph Matching Decoding (SMD) mechanism that exploits semantic relations among nodes to guide prediction. Furthermore, we design a Confusion-minimizing Node Contrast (CNC) loss to mitigate node ambiguity and subgraph heterogeneity by contrastively enhancing node discriminability in the graph space. Our method significantly outperforms prior CD-FSMIS approaches across multiple cross-domain benchmarks, achieving state-of-the-art performance while simultaneously preserving strong segmentation accuracy on the source domain.
Language as an Anchor: Preserving Relative Visual Geometry for Domain Incremental Learning
Nov 18, 2025Abstract:A key challenge in Domain Incremental Learning (DIL) is to continually learn under shifting distributions while preserving knowledge from previous domains. Existing methods face a fundamental dilemma. On one hand, projecting all domains into a single unified visual space leads to inter-domain interference and semantic distortion, as large shifts may vary with not only visual appearance but also underlying semantics. On the other hand, isolating domain-specific parameters causes knowledge fragmentation, creating "knowledge islands" that hamper knowledge reuse and exacerbate forgetting. To address this issue, we propose LAVA (Language-Anchored Visual Alignment), a novel DIL framework that replaces direct feature alignment with relative alignment driven by a text-based reference anchor. LAVA guides the visual representations of each incoming domain to preserve a consistent relative geometry, which is defined by mirroring the pairwise semantic similarities between the class names. This anchored geometric structure acts as a bridge across domains, enabling the retrieval of class-aware prior knowledge and facilitating robust feature aggregation. Extensive experiments on standard DIL benchmarks demonstrate that LAVA achieves significant performance improvements over state-of-the-arts. Code is available at https://github.com/ShuyiGeng/LAVA.
Hierarchical Spatio-temporal Segmentation Network for Ejection Fraction Estimation in Echocardiography Videos
Aug 26, 2025Abstract:Automated segmentation of the left ventricular endocardium in echocardiography videos is a key research area in cardiology. It aims to provide accurate assessment of cardiac structure and function through Ejection Fraction (EF) estimation. Although existing studies have achieved good segmentation performance, their results do not perform well in EF estimation. In this paper, we propose a Hierarchical Spatio-temporal Segmentation Network (\ourmodel) for echocardiography video, aiming to improve EF estimation accuracy by synergizing local detail modeling with global dynamic perception. The network employs a hierarchical design, with low-level stages using convolutional networks to process single-frame images and preserve details, while high-level stages utilize the Mamba architecture to capture spatio-temporal relationships. The hierarchical design balances single-frame and multi-frame processing, avoiding issues such as local error accumulation when relying solely on single frames or neglecting details when using only multi-frame data. To overcome local spatio-temporal limitations, we propose the Spatio-temporal Cross Scan (STCS) module, which integrates long-range context through skip scanning across frames and positions. This approach helps mitigate EF calculation biases caused by ultrasound image noise and other factors.
Frequency-adaptive tensor neural networks for high-dimensional multi-scale problems
Aug 21, 2025Abstract:Tensor neural networks (TNNs) have demonstrated their superiority in solving high-dimensional problems. However, similar to conventional neural networks, TNNs are also influenced by the Frequency Principle, which limits their ability to accurately capture high-frequency features of the solution. In this work, we analyze the training dynamics of TNNs by Fourier analysis and enhance their expressivity for high-dimensional multi-scale problems by incorporating random Fourier features. Leveraging the inherent tensor structure of TNNs, we further propose a novel approach to extract frequency features of high-dimensional functions by performing the Discrete Fourier Transform to one-dimensional component functions. This strategy effectively mitigates the curse of dimensionality. Building on this idea, we propose a frequency-adaptive TNNs algorithm, which significantly improves the ability of TNNs in solving complex multi-scale problems. Extensive numerical experiments are performed to validate the effectiveness and robustness of the proposed frequency-adaptive TNNs algorithm.
Uncertainty-aware Cross-training for Semi-supervised Medical Image Segmentation
Aug 12, 2025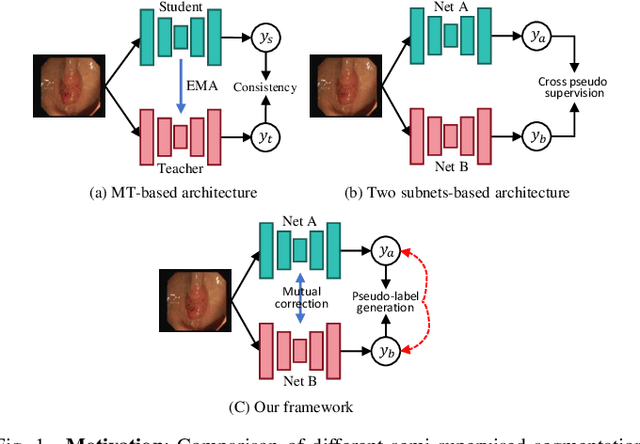
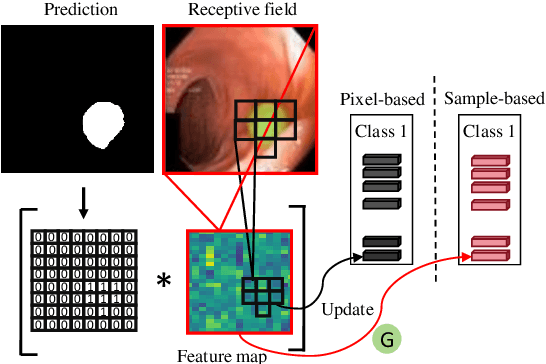
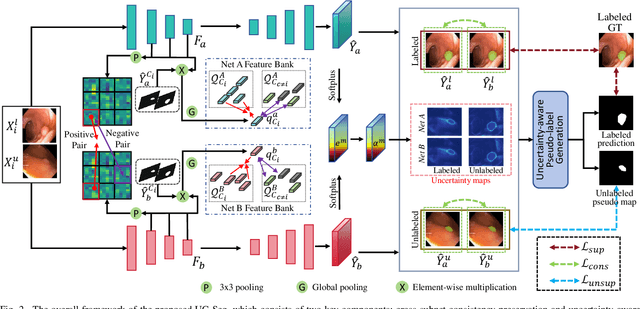
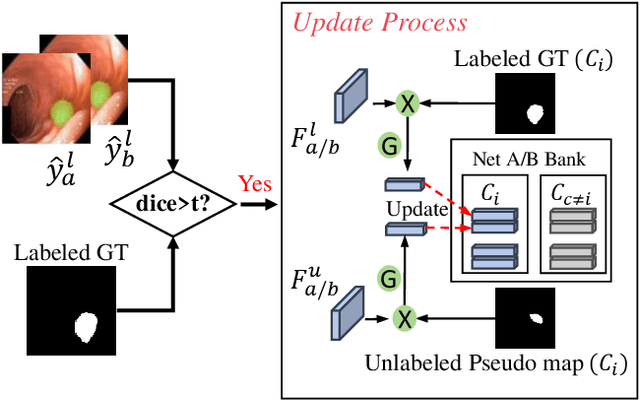
Abstract:Semi-supervised learning has gained considerable popularity in medical image segmentation tasks due to its capability to reduce reliance on expert-examined annotations. Several mean-teacher (MT) based semi-supervised methods utilize consistency regularization to effectively leverage valuable information from unlabeled data. However, these methods often heavily rely on the student model and overlook the potential impact of cognitive biases within the model. Furthermore, some methods employ co-training using pseudo-labels derived from different inputs, yet generating high-confidence pseudo-labels from perturbed inputs during training remains a significant challenge. In this paper, we propose an Uncertainty-aware Cross-training framework for semi-supervised medical image Segmentation (UC-Seg). Our UC-Seg framework incorporates two distinct subnets to effectively explore and leverage the correlation between them, thereby mitigating cognitive biases within the model. Specifically, we present a Cross-subnet Consistency Preservation (CCP) strategy to enhance feature representation capability and ensure feature consistency across the two subnets. This strategy enables each subnet to correct its own biases and learn shared semantics from both labeled and unlabeled data. Additionally, we propose an Uncertainty-aware Pseudo-label Generation (UPG) component that leverages segmentation results and corresponding uncertainty maps from both subnets to generate high-confidence pseudo-labels. We extensively evaluate the proposed UC-Seg on various medical image segmentation tasks involving different modality images, such as MRI, CT, ultrasound, colonoscopy, and so on. The results demonstrate that our method achieves superior segmentation accuracy and generalization performance compared to other state-of-the-art semi-supervised methods. Our code will be released at https://github.com/taozh2017/UCSeg.
LVM-GP: Uncertainty-Aware PDE Solver via coupling latent variable model and Gaussian process
Jul 30, 2025Abstract:We propose a novel probabilistic framework, termed LVM-GP, for uncertainty quantification in solving forward and inverse partial differential equations (PDEs) with noisy data. The core idea is to construct a stochastic mapping from the input to a high-dimensional latent representation, enabling uncertainty-aware prediction of the solution. Specifically, the architecture consists of a confidence-aware encoder and a probabilistic decoder. The encoder implements a high-dimensional latent variable model based on a Gaussian process (LVM-GP), where the latent representation is constructed by interpolating between a learnable deterministic feature and a Gaussian process prior, with the interpolation strength adaptively controlled by a confidence function learned from data. The decoder defines a conditional Gaussian distribution over the solution field, where the mean is predicted by a neural operator applied to the latent representation, allowing the model to learn flexible function-to-function mapping. Moreover, physical laws are enforced as soft constraints in the loss function to ensure consistency with the underlying PDE structure. Compared to existing approaches such as Bayesian physics-informed neural networks (B-PINNs) and deep ensembles, the proposed framework can efficiently capture functional dependencies via merging a latent Gaussian process and neural operator, resulting in competitive predictive accuracy and robust uncertainty quantification. Numerical experiments demonstrate the effectiveness and reliability of the method.
Text-driven Multiplanar Visual Interaction for Semi-supervised Medical Image Segmentation
Jul 16, 2025Abstract:Semi-supervised medical image segmentation is a crucial technique for alleviating the high cost of data annotation. When labeled data is limited, textual information can provide additional context to enhance visual semantic understanding. However, research exploring the use of textual data to enhance visual semantic embeddings in 3D medical imaging tasks remains scarce. In this paper, we propose a novel text-driven multiplanar visual interaction framework for semi-supervised medical image segmentation (termed Text-SemiSeg), which consists of three main modules: Text-enhanced Multiplanar Representation (TMR), Category-aware Semantic Alignment (CSA), and Dynamic Cognitive Augmentation (DCA). Specifically, TMR facilitates text-visual interaction through planar mapping, thereby enhancing the category awareness of visual features. CSA performs cross-modal semantic alignment between the text features with introduced learnable variables and the intermediate layer of visual features. DCA reduces the distribution discrepancy between labeled and unlabeled data through their interaction, thus improving the model's robustness. Finally, experiments on three public datasets demonstrate that our model effectively enhances visual features with textual information and outperforms other methods. Our code is available at https://github.com/taozh2017/Text-SemiSeg.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge