Huiqiao Xie
Limited-Angle CBCT Reconstruction via Geometry-Integrated Cycle-domain Denoising Diffusion Probabilistic Models
Jun 16, 2025



Abstract:Cone-beam CT (CBCT) is widely used in clinical radiotherapy for image-guided treatment, improving setup accuracy, adaptive planning, and motion management. However, slow gantry rotation limits performance by introducing motion artifacts, blurring, and increased dose. This work aims to develop a clinically feasible method for reconstructing high-quality CBCT volumes from consecutive limited-angle acquisitions, addressing imaging challenges in time- or dose-constrained settings. We propose a limited-angle (LA) geometry-integrated cycle-domain (LA-GICD) framework for CBCT reconstruction, comprising two denoising diffusion probabilistic models (DDPMs) connected via analytic cone-beam forward and back projectors. A Projection-DDPM completes missing projections, followed by back-projection, and an Image-DDPM refines the volume. This dual-domain design leverages complementary priors from projection and image spaces to achieve high-quality reconstructions from limited-angle (<= 90 degrees) scans. Performance was evaluated against full-angle reconstruction. Four board-certified medical physicists conducted assessments. A total of 78 planning CTs in common CBCT geometries were used for training and evaluation. The method achieved a mean absolute error of 35.5 HU, SSIM of 0.84, and PSNR of 29.8 dB, with visibly reduced artifacts and improved soft-tissue clarity. LA-GICD's geometry-aware dual-domain learning, embedded in analytic forward/backward operators, enabled artifact-free, high-contrast reconstructions from a single 90-degree scan, reducing acquisition time and dose four-fold. LA-GICD improves limited-angle CBCT reconstruction with strong data fidelity and anatomical realism. It offers a practical solution for short-arc acquisitions, enhancing CBCT use in radiotherapy by providing clinically applicable images with reduced scan time and dose for more accurate, personalized treatments.
Spatiotemporal Gaussian Optimization for 4D Cone Beam CT Reconstruction from Sparse Projections
Jan 07, 2025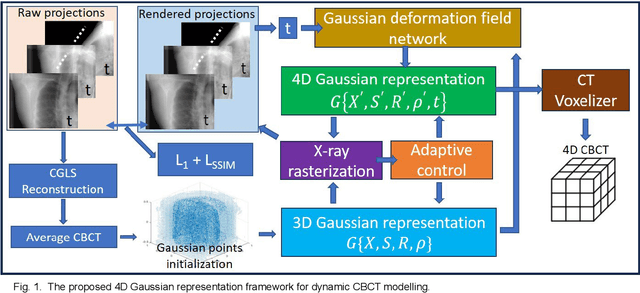
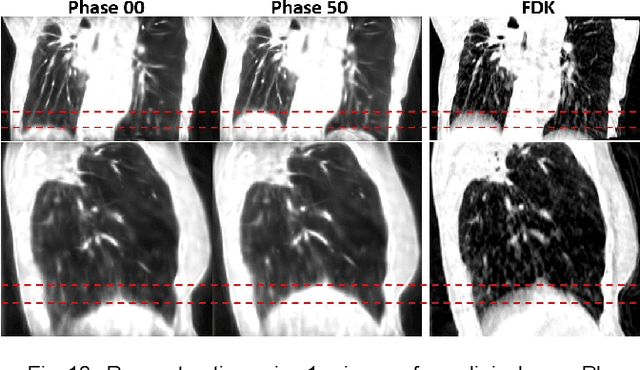
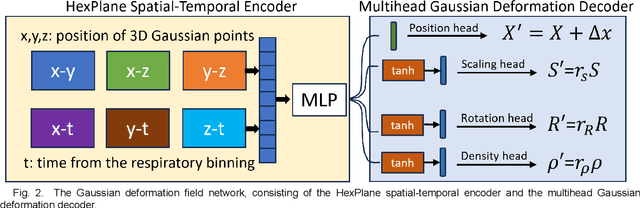
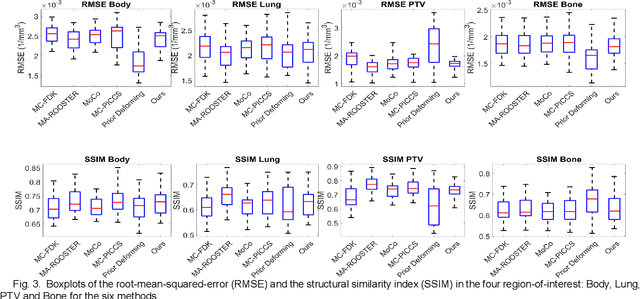
Abstract:In image-guided radiotherapy (IGRT), four-dimensional cone-beam computed tomography (4D-CBCT) is critical for assessing tumor motion during a patients breathing cycle prior to beam delivery. However, generating 4D-CBCT images with sufficient quality requires significantly more projection images than a standard 3D-CBCT scan, leading to extended scanning times and increased imaging dose to the patient. To address these limitations, there is a strong demand for methods capable of reconstructing high-quality 4D-CBCT images from a 1-minute 3D-CBCT acquisition. The challenge lies in the sparse sampling of projections, which introduces severe streaking artifacts and compromises image quality. This paper introduces a novel framework leveraging spatiotemporal Gaussian representation for 4D-CBCT reconstruction from sparse projections, achieving a balance between streak artifact reduction, dynamic motion preservation, and fine detail restoration. Each Gaussian is characterized by its 3D position, covariance, rotation, and density. Two-dimensional X-ray projection images can be rendered from the Gaussian point cloud representation via X-ray rasterization. The properties of each Gaussian were optimized by minimizing the discrepancy between the measured projections and the rendered X-ray projections. A Gaussian deformation network is jointly optimized to deform these Gaussian properties to obtain a 4D Gaussian representation for dynamic CBCT scene modeling. The final 4D-CBCT images are reconstructed by voxelizing the 4D Gaussians, achieving a high-quality representation that preserves both motion dynamics and spatial detail. The code and reconstruction results can be found at https://github.com/fuyabo/4DGS_for_4DCBCT/tree/main
Image-Domain Material Decomposition for Dual-energy CT using Unsupervised Learning with Data-fidelity Loss
Nov 17, 2023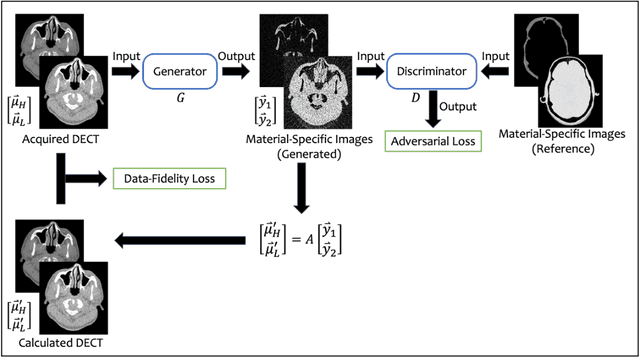
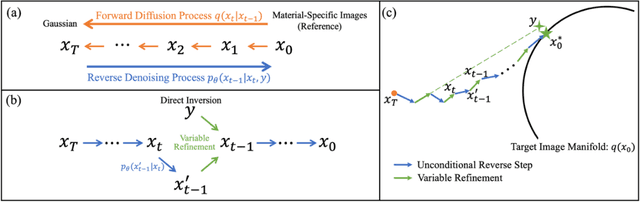
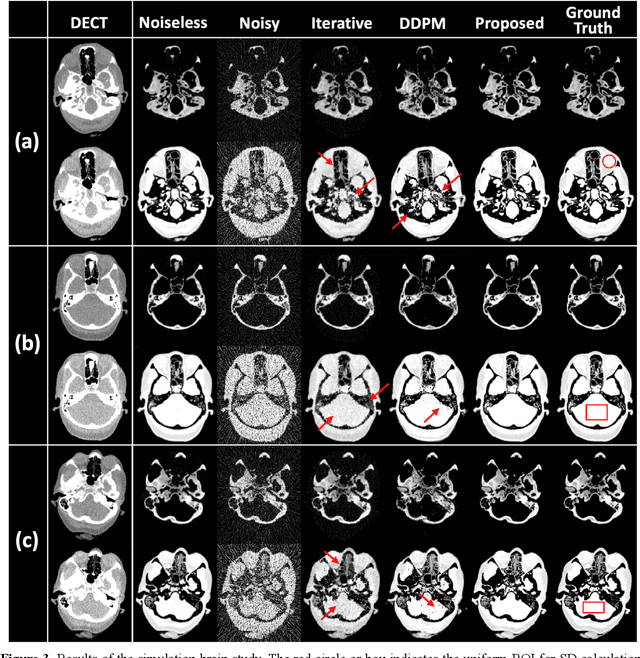
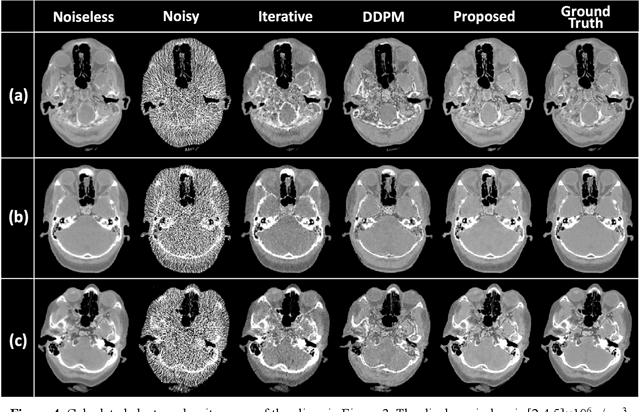
Abstract:Background: Dual-energy CT (DECT) and material decomposition play vital roles in quantitative medical imaging. However, the decomposition process may suffer from significant noise amplification, leading to severely degraded image signal-to-noise ratios (SNRs). While existing iterative algorithms perform noise suppression using different image priors, these heuristic image priors cannot accurately represent the features of the target image manifold. Although deep learning-based decomposition methods have been reported, these methods are in the supervised-learning framework requiring paired data for training, which is not readily available in clinical settings. Purpose: This work aims to develop an unsupervised-learning framework with data-measurement consistency for image-domain material decomposition in DECT.
Deformable Image Registration using Unsupervised Deep Learning for CBCT-guided Abdominal Radiotherapy
Aug 29, 2022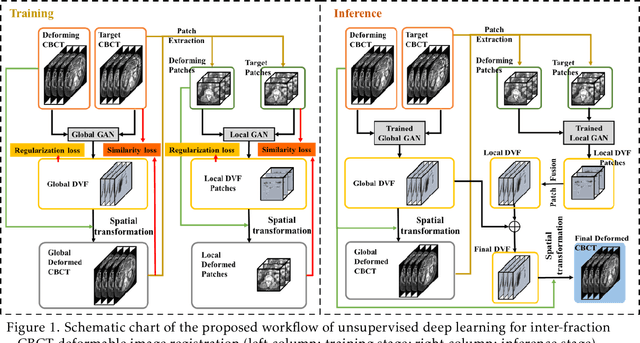
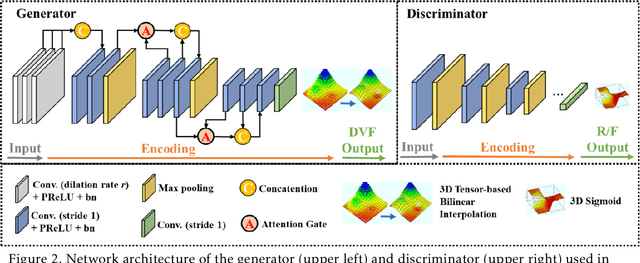
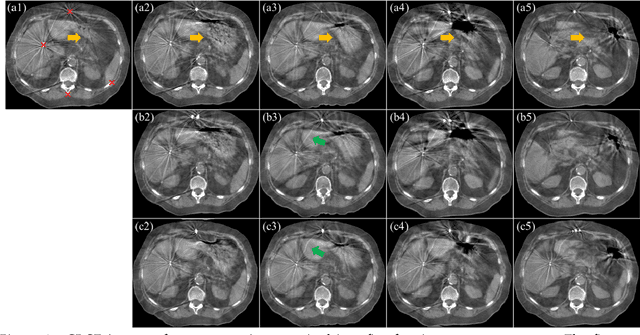
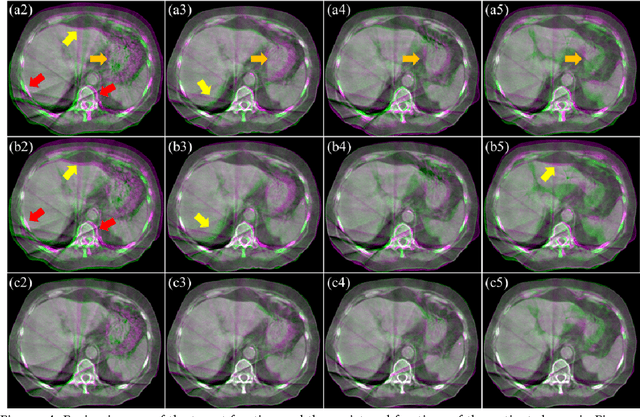
Abstract:CBCTs in image-guided radiotherapy provide crucial anatomy information for patient setup and plan evaluation. Longitudinal CBCT image registration could quantify the inter-fractional anatomic changes. The purpose of this study is to propose an unsupervised deep learning based CBCT-CBCT deformable image registration. The proposed deformable registration workflow consists of training and inference stages that share the same feed-forward path through a spatial transformation-based network (STN). The STN consists of a global generative adversarial network (GlobalGAN) and a local GAN (LocalGAN) to predict the coarse- and fine-scale motions, respectively. The network was trained by minimizing the image similarity loss and the deformable vector field (DVF) regularization loss without the supervision of ground truth DVFs. During the inference stage, patches of local DVF were predicted by the trained LocalGAN and fused to form a whole-image DVF. The local whole-image DVF was subsequently combined with the GlobalGAN generated DVF to obtain final DVF. The proposed method was evaluated using 100 fractional CBCTs from 20 abdominal cancer patients in the experiments and 105 fractional CBCTs from a cohort of 21 different abdominal cancer patients in a holdout test. Qualitatively, the registration results show great alignment between the deformed CBCT images and the target CBCT image. Quantitatively, the average target registration error (TRE) calculated on the fiducial markers and manually identified landmarks was 1.91+-1.11 mm. The average mean absolute error (MAE), normalized cross correlation (NCC) between the deformed CBCT and target CBCT were 33.42+-7.48 HU, 0.94+-0.04, respectively. This promising registration method could provide fast and accurate longitudinal CBCT alignment to facilitate inter-fractional anatomic changes analysis and prediction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge