Shaoyan Pan
Limited-Angle CBCT Reconstruction via Geometry-Integrated Cycle-domain Denoising Diffusion Probabilistic Models
Jun 16, 2025



Abstract:Cone-beam CT (CBCT) is widely used in clinical radiotherapy for image-guided treatment, improving setup accuracy, adaptive planning, and motion management. However, slow gantry rotation limits performance by introducing motion artifacts, blurring, and increased dose. This work aims to develop a clinically feasible method for reconstructing high-quality CBCT volumes from consecutive limited-angle acquisitions, addressing imaging challenges in time- or dose-constrained settings. We propose a limited-angle (LA) geometry-integrated cycle-domain (LA-GICD) framework for CBCT reconstruction, comprising two denoising diffusion probabilistic models (DDPMs) connected via analytic cone-beam forward and back projectors. A Projection-DDPM completes missing projections, followed by back-projection, and an Image-DDPM refines the volume. This dual-domain design leverages complementary priors from projection and image spaces to achieve high-quality reconstructions from limited-angle (<= 90 degrees) scans. Performance was evaluated against full-angle reconstruction. Four board-certified medical physicists conducted assessments. A total of 78 planning CTs in common CBCT geometries were used for training and evaluation. The method achieved a mean absolute error of 35.5 HU, SSIM of 0.84, and PSNR of 29.8 dB, with visibly reduced artifacts and improved soft-tissue clarity. LA-GICD's geometry-aware dual-domain learning, embedded in analytic forward/backward operators, enabled artifact-free, high-contrast reconstructions from a single 90-degree scan, reducing acquisition time and dose four-fold. LA-GICD improves limited-angle CBCT reconstruction with strong data fidelity and anatomical realism. It offers a practical solution for short-arc acquisitions, enhancing CBCT use in radiotherapy by providing clinically applicable images with reduced scan time and dose for more accurate, personalized treatments.
Label-Efficient Data Augmentation with Video Diffusion Models for Guidewire Segmentation in Cardiac Fluoroscopy
Dec 23, 2024Abstract:The accurate segmentation of guidewires in interventional cardiac fluoroscopy videos is crucial for computer-aided navigation tasks. Although deep learning methods have demonstrated high accuracy and robustness in wire segmentation, they require substantial annotated datasets for generalizability, underscoring the need for extensive labeled data to enhance model performance. To address this challenge, we propose the Segmentation-guided Frame-consistency Video Diffusion Model (SF-VD) to generate large collections of labeled fluoroscopy videos, augmenting the training data for wire segmentation networks. SF-VD leverages videos with limited annotations by independently modeling scene distribution and motion distribution. It first samples the scene distribution by generating 2D fluoroscopy images with wires positioned according to a specified input mask, and then samples the motion distribution by progressively generating subsequent frames, ensuring frame-to-frame coherence through a frame-consistency strategy. A segmentation-guided mechanism further refines the process by adjusting wire contrast, ensuring a diverse range of visibility in the synthesized image. Evaluation on a fluoroscopy dataset confirms the superior quality of the generated videos and shows significant improvements in guidewire segmentation.
AnatoMask: Enhancing Medical Image Segmentation with Reconstruction-guided Self-masking
Jul 09, 2024Abstract:Due to the scarcity of labeled data, self-supervised learning (SSL) has gained much attention in 3D medical image segmentation, by extracting semantic representations from unlabeled data. Among SSL strategies, Masked image modeling (MIM) has shown effectiveness by reconstructing randomly masked images to learn detailed representations. However, conventional MIM methods require extensive training data to achieve good performance, which still poses a challenge for medical imaging. Since random masking uniformly samples all regions within medical images, it may overlook crucial anatomical regions and thus degrade the pretraining efficiency. We propose AnatoMask, a novel MIM method that leverages reconstruction loss to dynamically identify and mask out anatomically significant regions to improve pretraining efficacy. AnatoMask takes a self-distillation approach, where the model learns both how to find more significant regions to mask and how to reconstruct these masked regions. To avoid suboptimal learning, Anatomask adjusts the pretraining difficulty progressively using a masking dynamics function. We have evaluated our method on 4 public datasets with multiple imaging modalities (CT, MRI, and PET). AnatoMask demonstrates superior performance and scalability compared to existing SSL methods. The code is available at https://github.com/ricklisz/AnatoMask.
Adaptive Self-Supervised Consistency-Guided Diffusion Model for Accelerated MRI Reconstruction
Jun 21, 2024Abstract:Purpose: To propose a self-supervised deep learning-based compressed sensing MRI (DL-based CS-MRI) method named "Adaptive Self-Supervised Consistency Guided Diffusion Model (ASSCGD)" to accelerate data acquisition without requiring fully sampled datasets. Materials and Methods: We used the fastMRI multi-coil brain axial T2-weighted (T2-w) dataset from 1,376 cases and single-coil brain quantitative magnetization prepared 2 rapid acquisition gradient echoes (MP2RAGE) T1 maps from 318 cases to train and test our model. Robustness against domain shift was evaluated using two out-of-distribution (OOD) datasets: multi-coil brain axial postcontrast T1 -weighted (T1c) dataset from 50 cases and axial T1-weighted (T1-w) dataset from 50 patients. Data were retrospectively subsampled at acceleration rates R in {2x, 4x, 8x}. ASSCGD partitions a random sampling pattern into two disjoint sets, ensuring data consistency during training. We compared our method with ReconFormer Transformer and SS-MRI, assessing performance using normalized mean squared error (NMSE), peak signal-to-noise ratio (PSNR), and structural similarity index (SSIM). Statistical tests included one-way analysis of variance (ANOVA) and multi-comparison Tukey's Honesty Significant Difference (HSD) tests. Results: ASSCGD preserved fine structures and brain abnormalities visually better than comparative methods at R = 8x for both multi-coil and single-coil datasets. It achieved the lowest NMSE at R in {4x, 8x}, and the highest PSNR and SSIM values at all acceleration rates for the multi-coil dataset. Similar trends were observed for the single-coil dataset, though SSIM values were comparable to ReconFormer at R in {2x, 8x}. These results were further confirmed by the voxel-wise correlation scatter plots. OOD results showed significant (p << 10^-5 ) improvements in undersampled image quality after reconstruction.
Clinically Feasible Diffusion Reconstruction for Highly-Accelerated Cardiac Cine MRI
Mar 13, 2024Abstract:The currently limited quality of accelerated cardiac cine reconstruction may potentially be improved by the emerging diffusion models, but the clinically unacceptable long processing time poses a challenge. We aim to develop a clinically feasible diffusion-model-based reconstruction pipeline to improve the image quality of cine MRI. A multi-in multi-out diffusion enhancement model together with fast inference strategies were developed to be used in conjunction with a reconstruction model. The diffusion reconstruction reduced spatial and temporal blurring in prospectively undersampled clinical data, as validated by experts inspection. The 1.5s per video processing time enabled the approach to be applied in clinical scenarios.
Spatiotemporal Diffusion Model with Paired Sampling for Accelerated Cardiac Cine MRI
Mar 13, 2024Abstract:Current deep learning reconstruction for accelerated cardiac cine MRI suffers from spatial and temporal blurring. We aim to improve image sharpness and motion delineation for cine MRI under high undersampling rates. A spatiotemporal diffusion enhancement model conditional on an existing deep learning reconstruction along with a novel paired sampling strategy was developed. The diffusion model provided sharper tissue boundaries and clearer motion than the original reconstruction in experts evaluation on clinical data. The innovative paired sampling strategy substantially reduced artificial noises in the generative results.
Multi-dimension unified Swin Transformer for 3D Lesion Segmentation in Multiple Anatomical Locations
Sep 04, 2023Abstract:In oncology research, accurate 3D segmentation of lesions from CT scans is essential for the modeling of lesion growth kinetics. However, following the RECIST criteria, radiologists routinely only delineate each lesion on the axial slice showing the largest transverse area, and delineate a small number of lesions in 3D for research purposes. As a result, we have plenty of unlabeled 3D volumes and labeled 2D images, and scarce labeled 3D volumes, which makes training a deep-learning 3D segmentation model a challenging task. In this work, we propose a novel model, denoted a multi-dimension unified Swin transformer (MDU-ST), for 3D lesion segmentation. The MDU-ST consists of a Shifted-window transformer (Swin-transformer) encoder and a convolutional neural network (CNN) decoder, allowing it to adapt to 2D and 3D inputs and learn the corresponding semantic information in the same encoder. Based on this model, we introduce a three-stage framework: 1) leveraging large amount of unlabeled 3D lesion volumes through self-supervised pretext tasks to learn the underlying pattern of lesion anatomy in the Swin-transformer encoder; 2) fine-tune the Swin-transformer encoder to perform 2D lesion segmentation with 2D RECIST slices to learn slice-level segmentation information; 3) further fine-tune the Swin-transformer encoder to perform 3D lesion segmentation with labeled 3D volumes. The network's performance is evaluated by the Dice similarity coefficient (DSC) and Hausdorff distance (HD) using an internal 3D lesion dataset with 593 lesions extracted from multiple anatomical locations. The proposed MDU-ST demonstrates significant improvement over the competing models. The proposed method can be used to conduct automated 3D lesion segmentation to assist radiomics and tumor growth modeling studies. This paper has been accepted by the IEEE International Symposium on Biomedical Imaging (ISBI) 2023.
Full-dose PET Synthesis from Low-dose PET Using High-efficiency Diffusion Denoising Probabilistic Model
Aug 24, 2023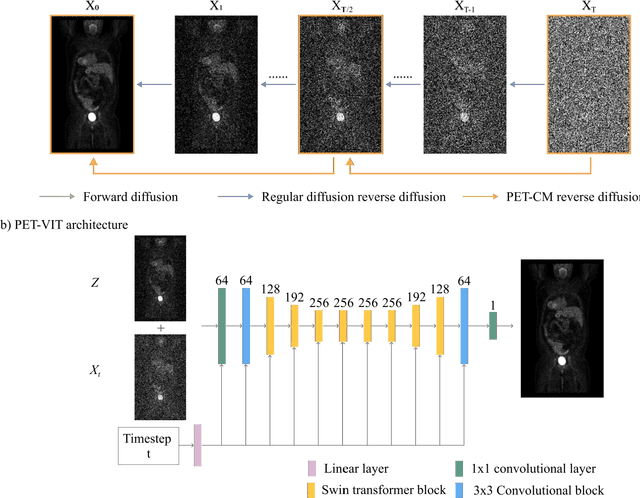


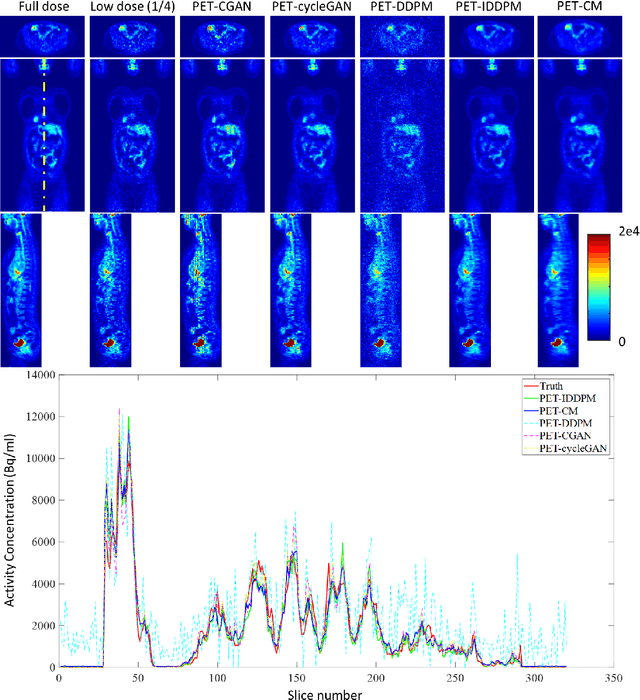
Abstract:To reduce the risks associated with ionizing radiation, a reduction of radiation exposure in PET imaging is needed. However, this leads to a detrimental effect on image contrast and quantification. High-quality PET images synthesized from low-dose data offer a solution to reduce radiation exposure. We introduce a diffusion-model-based approach for estimating full-dose PET images from low-dose ones: the PET Consistency Model (PET-CM) yielding synthetic quality comparable to state-of-the-art diffusion-based synthesis models, but with greater efficiency. There are two steps: a forward process that adds Gaussian noise to a full dose PET image at multiple timesteps, and a reverse diffusion process that employs a PET Shifted-window Vision Transformer (PET-VIT) network to learn the denoising procedure conditioned on the corresponding low-dose PETs. In PET-CM, the reverse process learns a consistency function for direct denoising of Gaussian noise to a clean full-dose PET. We evaluated the PET-CM in generating full-dose images using only 1/8 and 1/4 of the standard PET dose. Comparing 1/8 dose to full-dose images, PET-CM demonstrated impressive performance with normalized mean absolute error (NMAE) of 1.233+/-0.131%, peak signal-to-noise ratio (PSNR) of 33.915+/-0.933dB, structural similarity index (SSIM) of 0.964+/-0.009, and normalized cross-correlation (NCC) of 0.968+/-0.011, with an average generation time of 62 seconds per patient. This is a significant improvement compared to the state-of-the-art diffusion-based model with PET-CM reaching this result 12x faster. In the 1/4 dose to full-dose image experiments, PET-CM is also competitive, achieving an NMAE 1.058+/-0.092%, PSNR of 35.548+/-0.805dB, SSIM of 0.978+/-0.005, and NCC 0.981+/-0.007 The results indicate promising low-dose PET image quality improvements for clinical applications.
Synthetic CT Generation from MRI using 3D Transformer-based Denoising Diffusion Model
May 31, 2023


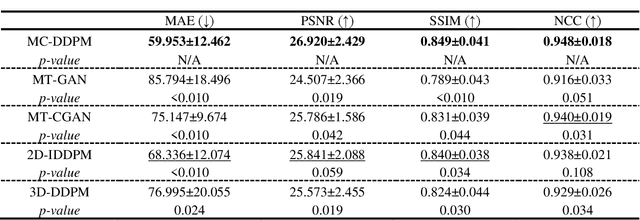
Abstract:Magnetic resonance imaging (MRI)-based synthetic computed tomography (sCT) simplifies radiation therapy treatment planning by eliminating the need for CT simulation and error-prone image registration, ultimately reducing patient radiation dose and setup uncertainty. We propose an MRI-to-CT transformer-based denoising diffusion probabilistic model (MC-DDPM) to transform MRI into high-quality sCT to facilitate radiation treatment planning. MC-DDPM implements diffusion processes with a shifted-window transformer network to generate sCT from MRI. The proposed model consists of two processes: a forward process which adds Gaussian noise to real CT scans, and a reverse process in which a shifted-window transformer V-net (Swin-Vnet) denoises the noisy CT scans conditioned on the MRI from the same patient to produce noise-free CT scans. With an optimally trained Swin-Vnet, the reverse diffusion process was used to generate sCT scans matching MRI anatomy. We evaluated the proposed method by generating sCT from MRI on a brain dataset and a prostate dataset. Qualitative evaluation was performed using the mean absolute error (MAE) of Hounsfield unit (HU), peak signal to noise ratio (PSNR), multi-scale Structure Similarity index (MS-SSIM) and normalized cross correlation (NCC) indexes between ground truth CTs and sCTs. MC-DDPM generated brain sCTs with state-of-the-art quantitative results with MAE 43.317 HU, PSNR 27.046 dB, SSIM 0.965, and NCC 0.983. For the prostate dataset, MC-DDPM achieved MAE 59.953 HU, PSNR 26.920 dB, SSIM 0.849, and NCC 0.948. In conclusion, we have developed and validated a novel approach for generating CT images from routine MRIs using a transformer-based DDPM. This model effectively captures the complex relationship between CT and MRI images, allowing for robust and high-quality synthetic CT (sCT) images to be generated in minutes.
Cross-Shaped Windows Transformer with Self-supervised Pretraining for Clinically Significant Prostate Cancer Detection in Bi-parametric MRI
Apr 30, 2023



Abstract:Multiparametric magnetic resonance imaging (mpMRI) has demonstrated promising results in prostate cancer (PCa) detection using deep convolutional neural networks (CNNs). Recently, transformers have achieved competitive performance compared to CNNs in computer vision. Large-scale transformers need abundant annotated data for training, which are difficult to obtain in medical imaging. Self-supervised learning can effectively leverage unlabeled data to extract useful semantic representations without annotation and its associated costs. This can improve model performance on downstream tasks with limited labelled data and increase generalizability. We introduce a novel end-to-end Cross-Shaped windows (CSwin) transformer UNet model, CSwin UNet, to detect clinically significant prostate cancer (csPCa) in prostate bi-parametric MR imaging (bpMRI) and demonstrate the effectiveness of our proposed self-supervised pre-training framework. Using a large prostate bpMRI dataset with 1500 patients, we first pre-train CSwin transformer using multi-task self-supervised learning to improve data-efficiency and network generalizability. We then finetuned using lesion annotations to perform csPCa detection. Five-fold cross validation shows that self-supervised CSwin UNet achieves 0.888 AUC and 0.545 Average Precision (AP), significantly outperforming four state-of-the-art models (Swin UNETR, DynUNet, Attention UNet, UNet). Using a separate bpMRI dataset with 158 patients, we evaluated our model robustness to external hold-out data. Self-supervised CSwin UNet achieves 0.79 AUC and 0.45 AP, still outperforming all other comparable methods and demonstrating generalization to a dataset shift.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge