Zach Eidex
Efficient Vision Mamba for MRI Super-Resolution via Hybrid Selective Scanning
Dec 22, 2025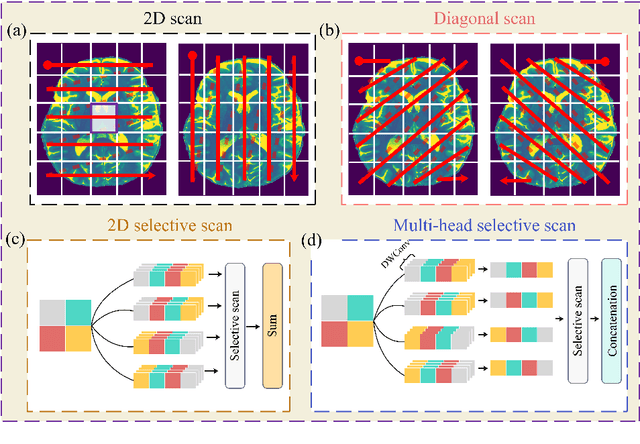
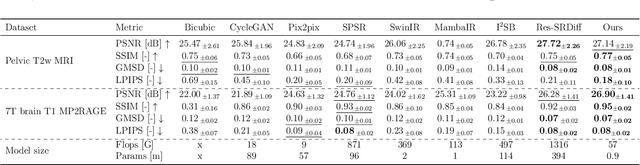
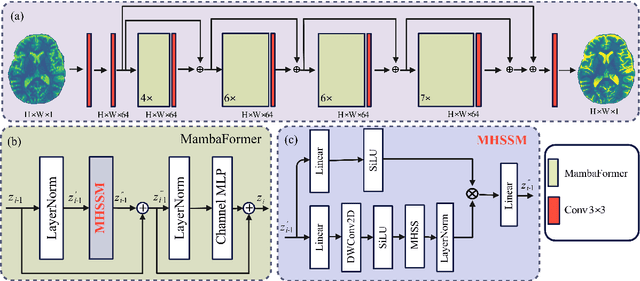

Abstract:Background: High-resolution MRI is critical for diagnosis, but long acquisition times limit clinical use. Super-resolution (SR) can enhance resolution post-scan, yet existing deep learning methods face fidelity-efficiency trade-offs. Purpose: To develop a computationally efficient and accurate deep learning framework for MRI SR that preserves anatomical detail for clinical integration. Materials and Methods: We propose a novel SR framework combining multi-head selective state-space models (MHSSM) with a lightweight channel MLP. The model uses 2D patch extraction with hybrid scanning to capture long-range dependencies. Each MambaFormer block integrates MHSSM, depthwise convolutions, and gated channel mixing. Evaluation used 7T brain T1 MP2RAGE maps (n=142) and 1.5T prostate T2w MRI (n=334). Comparisons included Bicubic interpolation, GANs (CycleGAN, Pix2pix, SPSR), transformers (SwinIR), Mamba (MambaIR), and diffusion models (I2SB, Res-SRDiff). Results: Our model achieved superior performance with exceptional efficiency. For 7T brain data: SSIM=0.951+-0.021, PSNR=26.90+-1.41 dB, LPIPS=0.076+-0.022, GMSD=0.083+-0.017, significantly outperforming all baselines (p<0.001). For prostate data: SSIM=0.770+-0.049, PSNR=27.15+-2.19 dB, LPIPS=0.190+-0.095, GMSD=0.087+-0.013. The framework used only 0.9M parameters and 57 GFLOPs, reducing parameters by 99.8% and computation by 97.5% versus Res-SRDiff, while outperforming SwinIR and MambaIR in accuracy and efficiency. Conclusion: The proposed framework provides an efficient, accurate MRI SR solution, delivering enhanced anatomical detail across datasets. Its low computational demand and state-of-the-art performance show strong potential for clinical translation.
Systematic Review and Meta-analysis of AI-driven MRI Motion Artifact Detection and Correction
Sep 05, 2025Abstract:Background: To systematically review and perform a meta-analysis of artificial intelligence (AI)-driven methods for detecting and correcting magnetic resonance imaging (MRI) motion artifacts, assessing current developments, effectiveness, challenges, and future research directions. Methods: A comprehensive systematic review and meta-analysis were conducted, focusing on deep learning (DL) approaches, particularly generative models, for the detection and correction of MRI motion artifacts. Quantitative data were extracted regarding utilized datasets, DL architectures, and performance metrics. Results: DL, particularly generative models, show promise for reducing motion artifacts and improving image quality; however, limited generalizability, reliance on paired training data, and risk of visual distortions remain key challenges that motivate standardized datasets and reporting. Conclusions: AI-driven methods, particularly DL generative models, show significant potential for improving MRI image quality by effectively addressing motion artifacts. However, critical challenges must be addressed, including the need for comprehensive public datasets, standardized reporting protocols for artifact levels, and more advanced, adaptable DL techniques to reduce reliance on extensive paired datasets. Addressing these aspects could substantially enhance MRI diagnostic accuracy, reduce healthcare costs, and improve patient care outcomes.
MRI motion correction via efficient residual-guided denoising diffusion probabilistic models
May 06, 2025Abstract:Purpose: Motion artifacts in magnetic resonance imaging (MRI) significantly degrade image quality and impair quantitative analysis. Conventional mitigation strategies, such as repeated acquisitions or motion tracking, are costly and workflow-intensive. This study introduces Res-MoCoDiff, an efficient denoising diffusion probabilistic model tailored for MRI motion artifact correction. Methods: Res-MoCoDiff incorporates a novel residual error shifting mechanism in the forward diffusion process, aligning the noise distribution with motion-corrupted data and enabling an efficient four-step reverse diffusion. A U-net backbone enhanced with Swin-Transformer blocks conventional attention layers, improving adaptability across resolutions. Training employs a combined l1+l2 loss, which promotes image sharpness and reduces pixel-level errors. Res-MoCoDiff was evaluated on synthetic dataset generated using a realistic motion simulation framework and on an in-vivo dataset. Comparative analyses were conducted against established methods, including CycleGAN, Pix2pix, and MT-DDPM using quantitative metrics such as peak signal-to-noise ratio (PSNR), structural similarity index measure (SSIM), and normalized mean squared error (NMSE). Results: The proposed method demonstrated superior performance in removing motion artifacts across all motion severity levels. Res-MoCoDiff consistently achieved the highest SSIM and the lowest NMSE values, with a PSNR of up to 41.91+-2.94 dB for minor distortions. Notably, the average sampling time was reduced to 0.37 seconds per batch of two image slices, compared with 101.74 seconds for conventional approaches.
MRI super-resolution reconstruction using efficient diffusion probabilistic model with residual shifting
Mar 03, 2025Abstract:Objective:This study introduces a residual error-shifting mechanism that drastically reduces sampling steps while preserving critical anatomical details, thus accelerating MRI reconstruction. Approach:We propose a novel diffusion-based SR framework called Res-SRDiff, which integrates residual error shifting into the forward diffusion process. This enables efficient HR image reconstruction by aligning the degraded HR and LR distributions.We evaluated Res-SRDiff on ultra-high-field brain T1 MP2RAGE maps and T2-weighted prostate images, comparing it with Bicubic, Pix2pix, CycleGAN, and a conventional denoising diffusion probabilistic model with vision transformer backbone (TM-DDPM), using quantitative metrics such as peak signal-to-noise ratio (PSNR), structural similarity index (SSIM), gradient magnitude similarity deviation (GMSD), and learned perceptual image patch similarity (LPIPS). Main results: Res-SRDiff significantly outperformed all comparative methods in terms of PSNR, SSIM, and GMSD across both datasets, with statistically significant improvements (p-values<<0.05). The model achieved high-fidelity image restoration with only four sampling steps, drastically reducing computational time to under one second per slice, which is substantially faster than conventional TM-DDPM with around 20 seconds per slice. Qualitative analyses further demonstrated that Res-SRDiff effectively preserved fine anatomical details and lesion morphology in both brain and pelvic MRI images. Significance: Our findings show that Res-SRDiff is an efficient and accurate MRI SR method, markedly improving computational efficiency and image quality. Integrating residual error shifting into the diffusion process allows for rapid and robust HR image reconstruction, enhancing clinical MRI workflows and advancing medical imaging research. The source at:https://github.com/mosaf/Res-SRDiff
A Physics-Informed Deep Learning Model for MRI Brain Motion Correction
Feb 13, 2025Abstract:Background: MRI is crucial for brain imaging but is highly susceptible to motion artifacts due to long acquisition times. This study introduces PI-MoCoNet, a physics-informed motion correction network that integrates spatial and k-space information to remove motion artifacts without explicit motion parameter estimation, enhancing image fidelity and diagnostic reliability. Materials and Methods: PI-MoCoNet consists of a motion detection network (U-net with spatial averaging) to identify corrupted k-space lines and a motion correction network (U-net with Swin Transformer blocks) to reconstruct motion-free images. The correction is guided by three loss functions: reconstruction (L1), perceptual (LPIPS), and data consistency (Ldc). Motion artifacts were simulated via rigid phase encoding perturbations and evaluated on IXI and MR-ART datasets against Pix2Pix, CycleGAN, and U-net using PSNR, SSIM, and NMSE. Results: PI-MoCoNet significantly improved image quality. On IXI, for minor artifacts, PSNR increased from 34.15 dB to 45.95 dB, SSIM from 0.87 to 1.00, and NMSE reduced from 0.55% to 0.04%. For moderate artifacts, PSNR improved from 30.23 dB to 42.16 dB, SSIM from 0.80 to 0.99, and NMSE from 1.32% to 0.09%. For heavy artifacts, PSNR rose from 27.99 dB to 36.01 dB, SSIM from 0.75 to 0.97, and NMSE decreased from 2.21% to 0.36%. On MR-ART, PI-MoCoNet achieved PSNR gains of ~10 dB and SSIM improvements of up to 0.20, with NMSE reductions of ~6%. Ablation studies confirmed the importance of data consistency and perceptual losses, yielding a 1 dB PSNR gain and 0.17% NMSE reduction. Conclusions: PI-MoCoNet effectively mitigates motion artifacts in brain MRI, outperforming existing methods. Its ability to integrate spatial and k-space information makes it a promising tool for clinical use in motion-prone settings. Code: https://github.com/mosaf/PI-MoCoNet.git.
Advancing MRI Reconstruction: A Systematic Review of Deep Learning and Compressed Sensing Integration
Jan 24, 2025
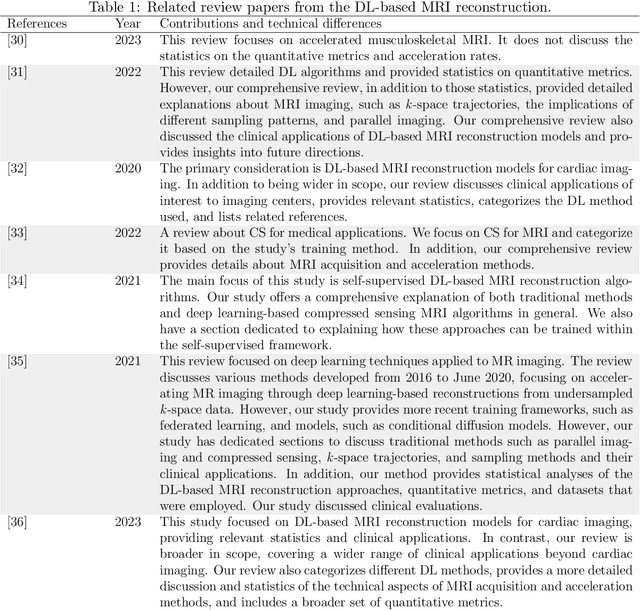
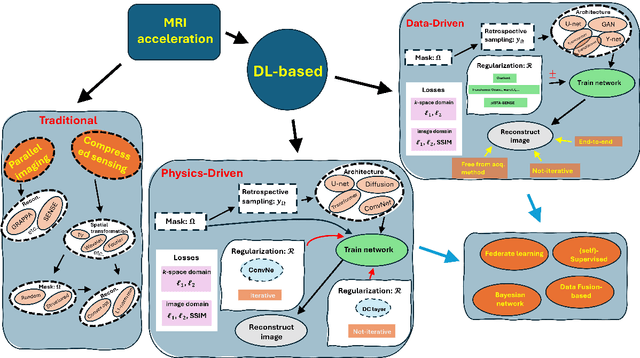
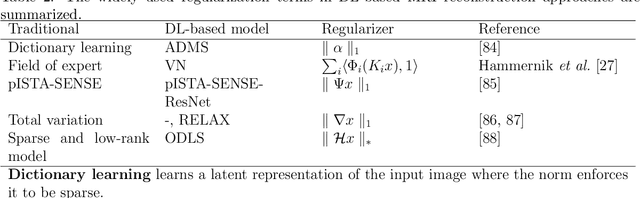
Abstract:Magnetic resonance imaging (MRI) is a non-invasive imaging modality and provides comprehensive anatomical and functional insights into the human body. However, its long acquisition times can lead to patient discomfort, motion artifacts, and limiting real-time applications. To address these challenges, strategies such as parallel imaging have been applied, which utilize multiple receiver coils to speed up the data acquisition process. Additionally, compressed sensing (CS) is a method that facilitates image reconstruction from sparse data, significantly reducing image acquisition time by minimizing the amount of data collection needed. Recently, deep learning (DL) has emerged as a powerful tool for improving MRI reconstruction. It has been integrated with parallel imaging and CS principles to achieve faster and more accurate MRI reconstructions. This review comprehensively examines DL-based techniques for MRI reconstruction. We categorize and discuss various DL-based methods, including end-to-end approaches, unrolled optimization, and federated learning, highlighting their potential benefits. Our systematic review highlights significant contributions and underscores the potential of DL in MRI reconstruction. Additionally, we summarize key results and trends in DL-based MRI reconstruction, including quantitative metrics, the dataset, acceleration factors, and the progress of and research interest in DL techniques over time. Finally, we discuss potential future directions and the importance of DL-based MRI reconstruction in advancing medical imaging. To facilitate further research in this area, we provide a GitHub repository that includes up-to-date DL-based MRI reconstruction publications and public datasets-https://github.com/mosaf/Awesome-DL-based-CS-MRI.
T1-contrast Enhanced MRI Generation from Multi-parametric MRI for Glioma Patients with Latent Tumor Conditioning
Sep 03, 2024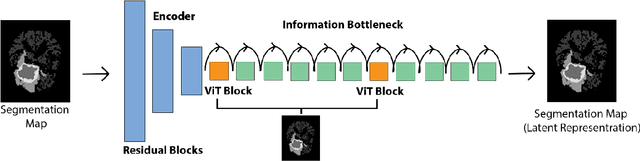
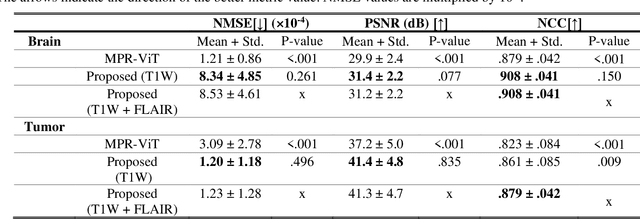
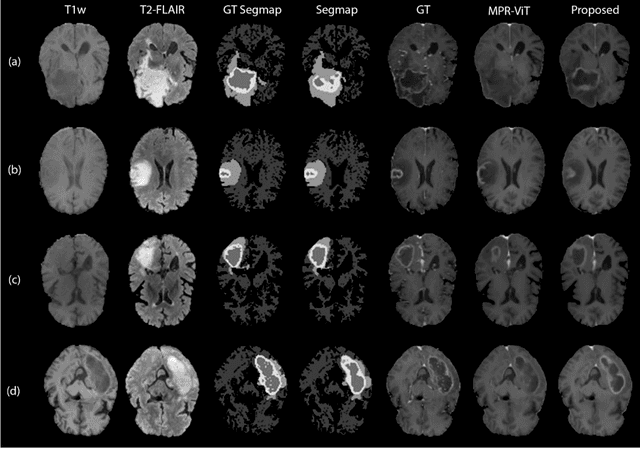
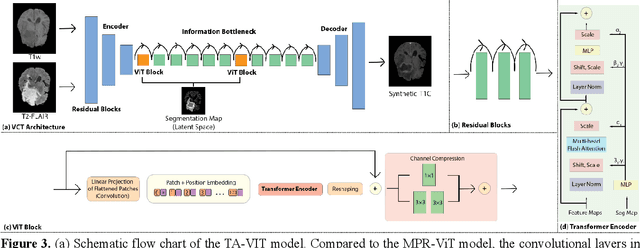
Abstract:Objective: Gadolinium-based contrast agents (GBCAs) are commonly used in MRI scans of patients with gliomas to enhance brain tumor characterization using T1-weighted (T1W) MRI. However, there is growing concern about GBCA toxicity. This study develops a deep-learning framework to generate T1-postcontrast (T1C) from pre-contrast multiparametric MRI. Approach: We propose the tumor-aware vision transformer (TA-ViT) model that predicts high-quality T1C images. The predicted tumor region is significantly improved (P < .001) by conditioning the transformer layers from predicted segmentation maps through adaptive layer norm zero mechanism. The predicted segmentation maps were generated with the multi-parametric residual (MPR) ViT model and transformed into a latent space to produce compressed, feature-rich representations. The TA-ViT model predicted T1C MRI images of 501 glioma cases. Selected patients were split into training (N=400), validation (N=50), and test (N=51) sets. Main Results: Both qualitative and quantitative results demonstrate that the TA-ViT model performs superior against the benchmark MRP-ViT model. Our method produces synthetic T1C MRI with high soft tissue contrast and more accurately reconstructs both the tumor and whole brain volumes. The synthesized T1C images achieved remarkable improvements in both tumor and healthy tissue regions compared to the MRP-ViT model. For healthy tissue and tumor regions, the results were as follows: NMSE: 8.53 +/- 4.61E-4; PSNR: 31.2 +/- 2.2; NCC: 0.908 +/- .041 and NMSE: 1.22 +/- 1.27E-4, PSNR: 41.3 +/- 4.7, and NCC: 0.879 +/- 0.042, respectively. Significance: The proposed method generates synthetic T1C images that closely resemble real T1C images. Future development and application of this approach may enable contrast-agent-free MRI for brain tumor patients, eliminating the risk of GBCA toxicity and simplifying the MRI scan protocol.
Deep Learning Based Apparent Diffusion Coefficient Map Generation1 from Multi-parametric MR Images for Patients with Diffuse Gliomas
Jul 02, 2024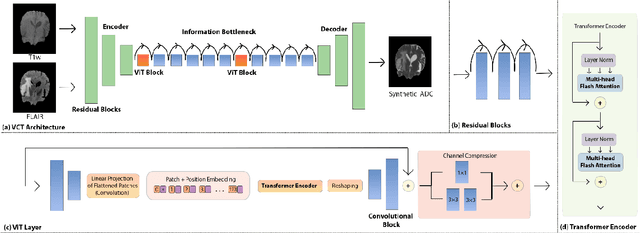
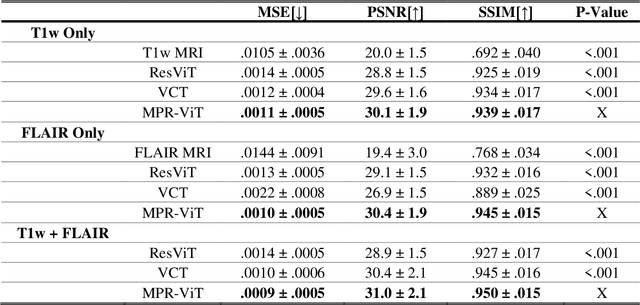
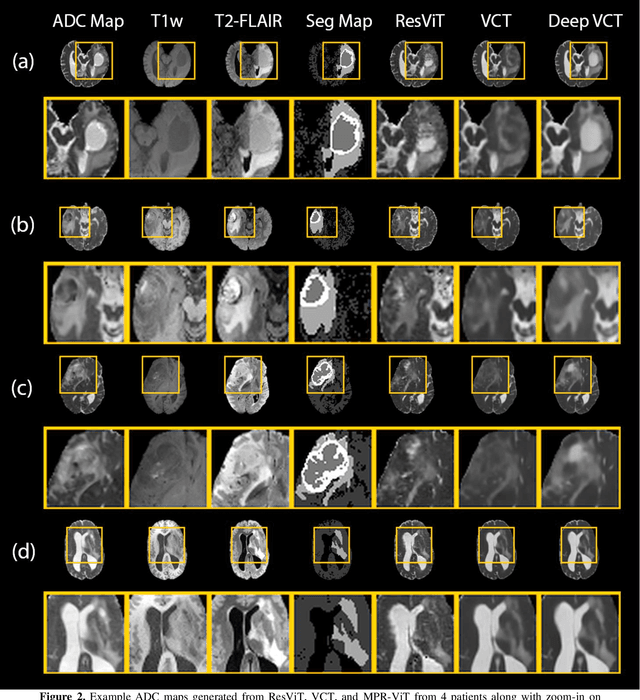
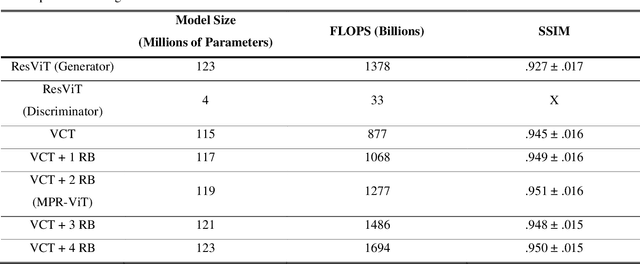
Abstract:Purpose: Apparent diffusion coefficient (ADC) maps derived from diffusion weighted (DWI) MRI provides functional measurements about the water molecules in tissues. However, DWI is time consuming and very susceptible to image artifacts, leading to inaccurate ADC measurements. This study aims to develop a deep learning framework to synthesize ADC maps from multi-parametric MR images. Methods: We proposed the multiparametric residual vision transformer model (MPR-ViT) that leverages the long-range context of ViT layers along with the precision of convolutional operators. Residual blocks throughout the network significantly increasing the representational power of the model. The MPR-ViT model was applied to T1w and T2- fluid attenuated inversion recovery images of 501 glioma cases from a publicly available dataset including preprocessed ADC maps. Selected patients were divided into training (N=400), validation (N=50) and test (N=51) sets, respectively. Using the preprocessed ADC maps as ground truth, model performance was evaluated and compared against the Vision Convolutional Transformer (VCT) and residual vision transformer (ResViT) models. Results: The results are as follows using T1w + T2-FLAIR MRI as inputs: MPR-ViT - PSNR: 31.0 +/- 2.1, MSE: 0.009 +/- 0.0005, SSIM: 0.950 +/- 0.015. In addition, ablation studies showed the relative impact on performance of each input sequence. Both qualitative and quantitative results indicate that the proposed MR- ViT model performs favorably against the ground truth data. Conclusion: We show that high-quality ADC maps can be synthesized from structural MRI using a MPR- VCT model. Our predicted images show better conformality to the ground truth volume than ResViT and VCT predictions. These high-quality synthetic ADC maps would be particularly useful for disease diagnosis and intervention, especially when ADC maps have artifacts or are unavailable.
Adaptive Self-Supervised Consistency-Guided Diffusion Model for Accelerated MRI Reconstruction
Jun 21, 2024Abstract:Purpose: To propose a self-supervised deep learning-based compressed sensing MRI (DL-based CS-MRI) method named "Adaptive Self-Supervised Consistency Guided Diffusion Model (ASSCGD)" to accelerate data acquisition without requiring fully sampled datasets. Materials and Methods: We used the fastMRI multi-coil brain axial T2-weighted (T2-w) dataset from 1,376 cases and single-coil brain quantitative magnetization prepared 2 rapid acquisition gradient echoes (MP2RAGE) T1 maps from 318 cases to train and test our model. Robustness against domain shift was evaluated using two out-of-distribution (OOD) datasets: multi-coil brain axial postcontrast T1 -weighted (T1c) dataset from 50 cases and axial T1-weighted (T1-w) dataset from 50 patients. Data were retrospectively subsampled at acceleration rates R in {2x, 4x, 8x}. ASSCGD partitions a random sampling pattern into two disjoint sets, ensuring data consistency during training. We compared our method with ReconFormer Transformer and SS-MRI, assessing performance using normalized mean squared error (NMSE), peak signal-to-noise ratio (PSNR), and structural similarity index (SSIM). Statistical tests included one-way analysis of variance (ANOVA) and multi-comparison Tukey's Honesty Significant Difference (HSD) tests. Results: ASSCGD preserved fine structures and brain abnormalities visually better than comparative methods at R = 8x for both multi-coil and single-coil datasets. It achieved the lowest NMSE at R in {4x, 8x}, and the highest PSNR and SSIM values at all acceleration rates for the multi-coil dataset. Similar trends were observed for the single-coil dataset, though SSIM values were comparable to ReconFormer at R in {2x, 8x}. These results were further confirmed by the voxel-wise correlation scatter plots. OOD results showed significant (p << 10^-5 ) improvements in undersampled image quality after reconstruction.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge