Hui Mao
Efficient Vision Mamba for MRI Super-Resolution via Hybrid Selective Scanning
Dec 22, 2025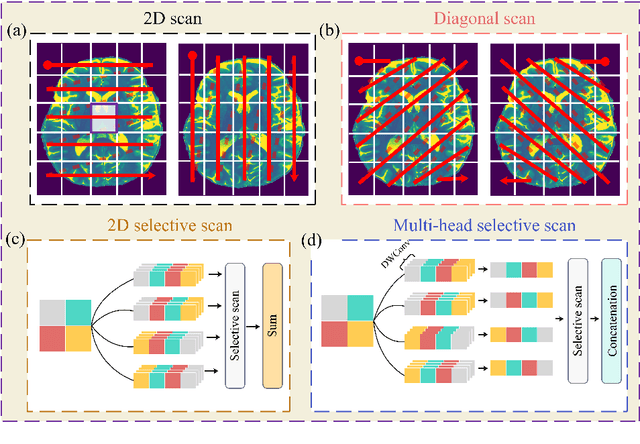
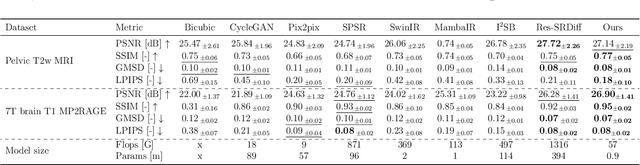
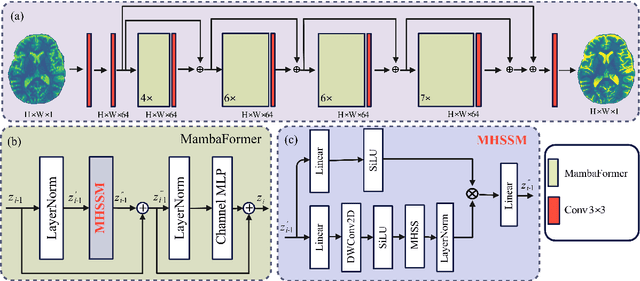

Abstract:Background: High-resolution MRI is critical for diagnosis, but long acquisition times limit clinical use. Super-resolution (SR) can enhance resolution post-scan, yet existing deep learning methods face fidelity-efficiency trade-offs. Purpose: To develop a computationally efficient and accurate deep learning framework for MRI SR that preserves anatomical detail for clinical integration. Materials and Methods: We propose a novel SR framework combining multi-head selective state-space models (MHSSM) with a lightweight channel MLP. The model uses 2D patch extraction with hybrid scanning to capture long-range dependencies. Each MambaFormer block integrates MHSSM, depthwise convolutions, and gated channel mixing. Evaluation used 7T brain T1 MP2RAGE maps (n=142) and 1.5T prostate T2w MRI (n=334). Comparisons included Bicubic interpolation, GANs (CycleGAN, Pix2pix, SPSR), transformers (SwinIR), Mamba (MambaIR), and diffusion models (I2SB, Res-SRDiff). Results: Our model achieved superior performance with exceptional efficiency. For 7T brain data: SSIM=0.951+-0.021, PSNR=26.90+-1.41 dB, LPIPS=0.076+-0.022, GMSD=0.083+-0.017, significantly outperforming all baselines (p<0.001). For prostate data: SSIM=0.770+-0.049, PSNR=27.15+-2.19 dB, LPIPS=0.190+-0.095, GMSD=0.087+-0.013. The framework used only 0.9M parameters and 57 GFLOPs, reducing parameters by 99.8% and computation by 97.5% versus Res-SRDiff, while outperforming SwinIR and MambaIR in accuracy and efficiency. Conclusion: The proposed framework provides an efficient, accurate MRI SR solution, delivering enhanced anatomical detail across datasets. Its low computational demand and state-of-the-art performance show strong potential for clinical translation.
MRI motion correction via efficient residual-guided denoising diffusion probabilistic models
May 06, 2025Abstract:Purpose: Motion artifacts in magnetic resonance imaging (MRI) significantly degrade image quality and impair quantitative analysis. Conventional mitigation strategies, such as repeated acquisitions or motion tracking, are costly and workflow-intensive. This study introduces Res-MoCoDiff, an efficient denoising diffusion probabilistic model tailored for MRI motion artifact correction. Methods: Res-MoCoDiff incorporates a novel residual error shifting mechanism in the forward diffusion process, aligning the noise distribution with motion-corrupted data and enabling an efficient four-step reverse diffusion. A U-net backbone enhanced with Swin-Transformer blocks conventional attention layers, improving adaptability across resolutions. Training employs a combined l1+l2 loss, which promotes image sharpness and reduces pixel-level errors. Res-MoCoDiff was evaluated on synthetic dataset generated using a realistic motion simulation framework and on an in-vivo dataset. Comparative analyses were conducted against established methods, including CycleGAN, Pix2pix, and MT-DDPM using quantitative metrics such as peak signal-to-noise ratio (PSNR), structural similarity index measure (SSIM), and normalized mean squared error (NMSE). Results: The proposed method demonstrated superior performance in removing motion artifacts across all motion severity levels. Res-MoCoDiff consistently achieved the highest SSIM and the lowest NMSE values, with a PSNR of up to 41.91+-2.94 dB for minor distortions. Notably, the average sampling time was reduced to 0.37 seconds per batch of two image slices, compared with 101.74 seconds for conventional approaches.
T1-contrast Enhanced MRI Generation from Multi-parametric MRI for Glioma Patients with Latent Tumor Conditioning
Sep 03, 2024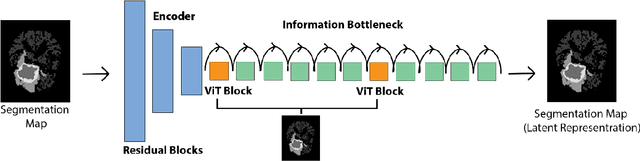
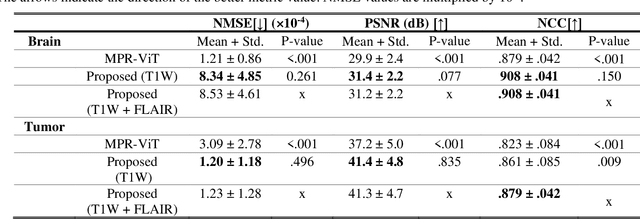
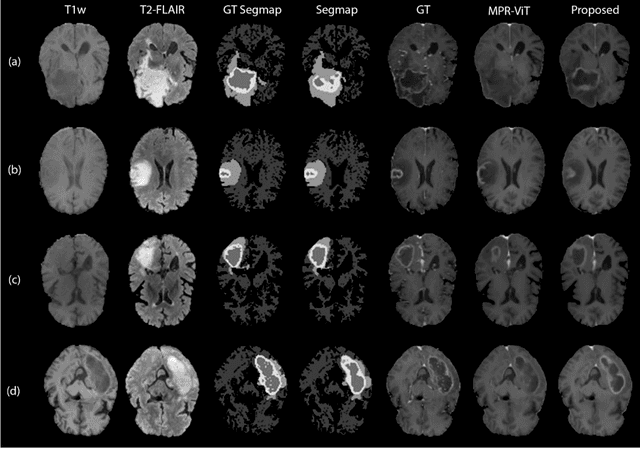
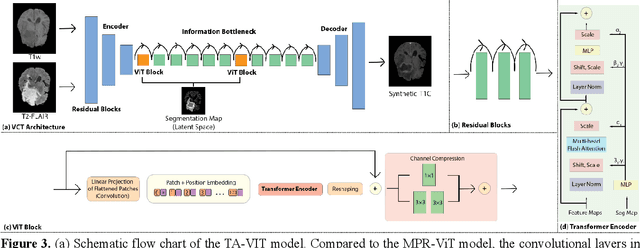
Abstract:Objective: Gadolinium-based contrast agents (GBCAs) are commonly used in MRI scans of patients with gliomas to enhance brain tumor characterization using T1-weighted (T1W) MRI. However, there is growing concern about GBCA toxicity. This study develops a deep-learning framework to generate T1-postcontrast (T1C) from pre-contrast multiparametric MRI. Approach: We propose the tumor-aware vision transformer (TA-ViT) model that predicts high-quality T1C images. The predicted tumor region is significantly improved (P < .001) by conditioning the transformer layers from predicted segmentation maps through adaptive layer norm zero mechanism. The predicted segmentation maps were generated with the multi-parametric residual (MPR) ViT model and transformed into a latent space to produce compressed, feature-rich representations. The TA-ViT model predicted T1C MRI images of 501 glioma cases. Selected patients were split into training (N=400), validation (N=50), and test (N=51) sets. Main Results: Both qualitative and quantitative results demonstrate that the TA-ViT model performs superior against the benchmark MRP-ViT model. Our method produces synthetic T1C MRI with high soft tissue contrast and more accurately reconstructs both the tumor and whole brain volumes. The synthesized T1C images achieved remarkable improvements in both tumor and healthy tissue regions compared to the MRP-ViT model. For healthy tissue and tumor regions, the results were as follows: NMSE: 8.53 +/- 4.61E-4; PSNR: 31.2 +/- 2.2; NCC: 0.908 +/- .041 and NMSE: 1.22 +/- 1.27E-4, PSNR: 41.3 +/- 4.7, and NCC: 0.879 +/- 0.042, respectively. Significance: The proposed method generates synthetic T1C images that closely resemble real T1C images. Future development and application of this approach may enable contrast-agent-free MRI for brain tumor patients, eliminating the risk of GBCA toxicity and simplifying the MRI scan protocol.
Deep Learning Based Apparent Diffusion Coefficient Map Generation1 from Multi-parametric MR Images for Patients with Diffuse Gliomas
Jul 02, 2024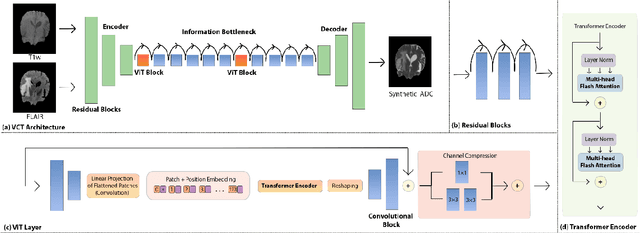
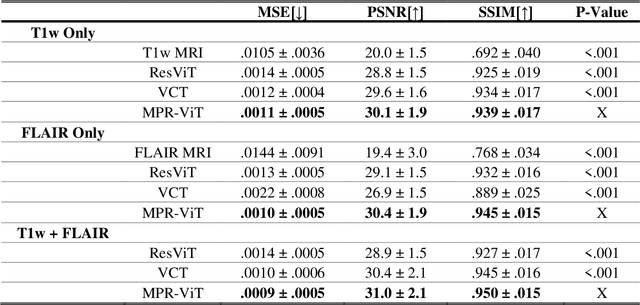
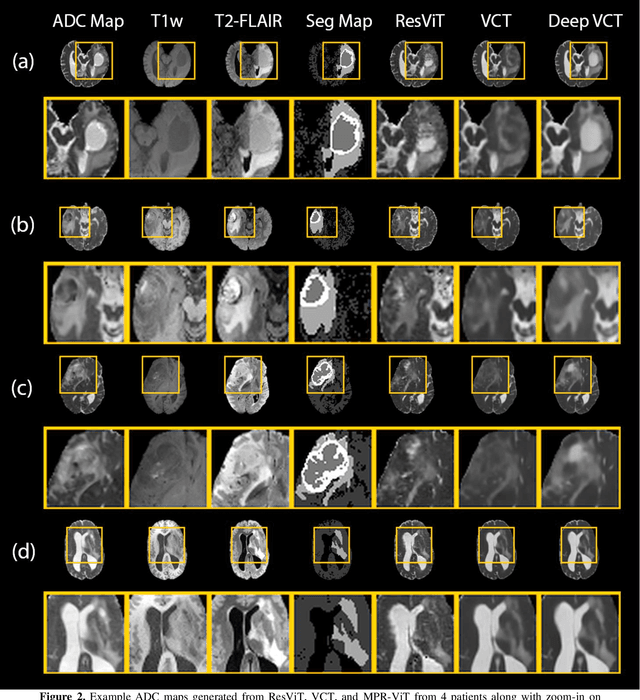
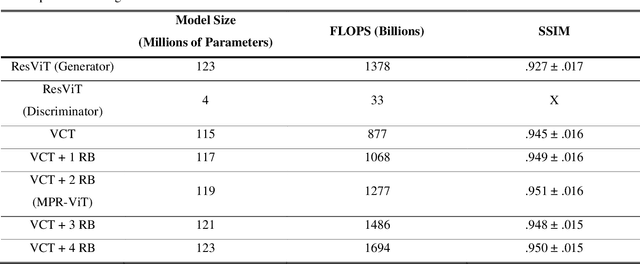
Abstract:Purpose: Apparent diffusion coefficient (ADC) maps derived from diffusion weighted (DWI) MRI provides functional measurements about the water molecules in tissues. However, DWI is time consuming and very susceptible to image artifacts, leading to inaccurate ADC measurements. This study aims to develop a deep learning framework to synthesize ADC maps from multi-parametric MR images. Methods: We proposed the multiparametric residual vision transformer model (MPR-ViT) that leverages the long-range context of ViT layers along with the precision of convolutional operators. Residual blocks throughout the network significantly increasing the representational power of the model. The MPR-ViT model was applied to T1w and T2- fluid attenuated inversion recovery images of 501 glioma cases from a publicly available dataset including preprocessed ADC maps. Selected patients were divided into training (N=400), validation (N=50) and test (N=51) sets, respectively. Using the preprocessed ADC maps as ground truth, model performance was evaluated and compared against the Vision Convolutional Transformer (VCT) and residual vision transformer (ResViT) models. Results: The results are as follows using T1w + T2-FLAIR MRI as inputs: MPR-ViT - PSNR: 31.0 +/- 2.1, MSE: 0.009 +/- 0.0005, SSIM: 0.950 +/- 0.015. In addition, ablation studies showed the relative impact on performance of each input sequence. Both qualitative and quantitative results indicate that the proposed MR- ViT model performs favorably against the ground truth data. Conclusion: We show that high-quality ADC maps can be synthesized from structural MRI using a MPR- VCT model. Our predicted images show better conformality to the ground truth volume than ResViT and VCT predictions. These high-quality synthetic ADC maps would be particularly useful for disease diagnosis and intervention, especially when ADC maps have artifacts or are unavailable.
Full-dose PET Synthesis from Low-dose PET Using High-efficiency Diffusion Denoising Probabilistic Model
Aug 24, 2023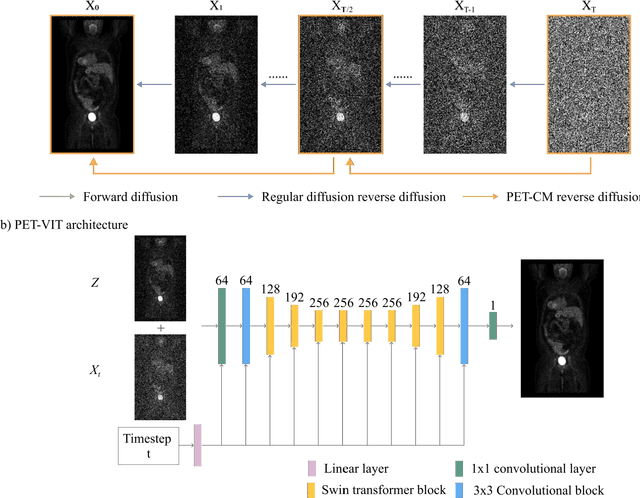


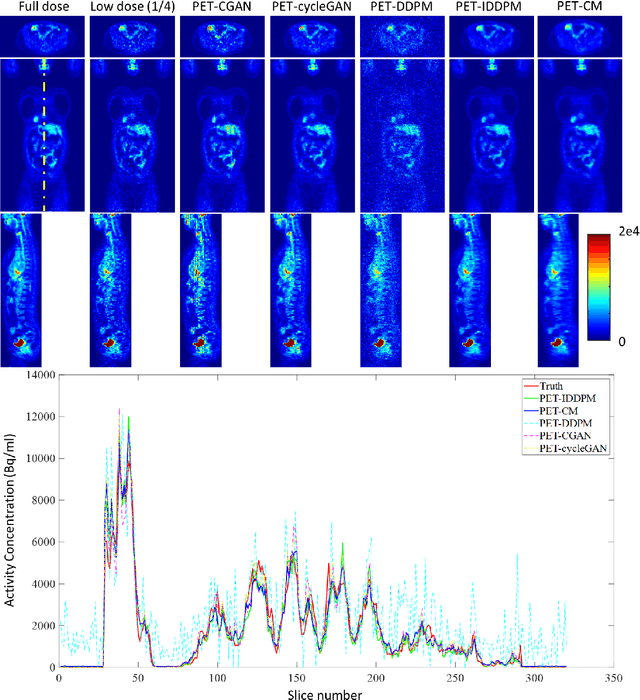
Abstract:To reduce the risks associated with ionizing radiation, a reduction of radiation exposure in PET imaging is needed. However, this leads to a detrimental effect on image contrast and quantification. High-quality PET images synthesized from low-dose data offer a solution to reduce radiation exposure. We introduce a diffusion-model-based approach for estimating full-dose PET images from low-dose ones: the PET Consistency Model (PET-CM) yielding synthetic quality comparable to state-of-the-art diffusion-based synthesis models, but with greater efficiency. There are two steps: a forward process that adds Gaussian noise to a full dose PET image at multiple timesteps, and a reverse diffusion process that employs a PET Shifted-window Vision Transformer (PET-VIT) network to learn the denoising procedure conditioned on the corresponding low-dose PETs. In PET-CM, the reverse process learns a consistency function for direct denoising of Gaussian noise to a clean full-dose PET. We evaluated the PET-CM in generating full-dose images using only 1/8 and 1/4 of the standard PET dose. Comparing 1/8 dose to full-dose images, PET-CM demonstrated impressive performance with normalized mean absolute error (NMAE) of 1.233+/-0.131%, peak signal-to-noise ratio (PSNR) of 33.915+/-0.933dB, structural similarity index (SSIM) of 0.964+/-0.009, and normalized cross-correlation (NCC) of 0.968+/-0.011, with an average generation time of 62 seconds per patient. This is a significant improvement compared to the state-of-the-art diffusion-based model with PET-CM reaching this result 12x faster. In the 1/4 dose to full-dose image experiments, PET-CM is also competitive, achieving an NMAE 1.058+/-0.092%, PSNR of 35.548+/-0.805dB, SSIM of 0.978+/-0.005, and NCC 0.981+/-0.007 The results indicate promising low-dose PET image quality improvements for clinical applications.
Synthetic CT Generation from MRI using 3D Transformer-based Denoising Diffusion Model
May 31, 2023


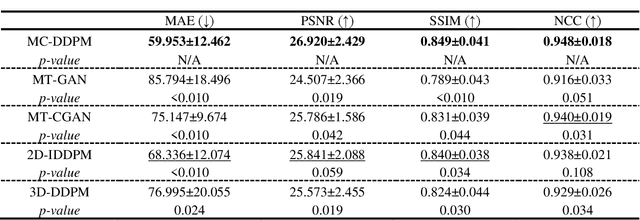
Abstract:Magnetic resonance imaging (MRI)-based synthetic computed tomography (sCT) simplifies radiation therapy treatment planning by eliminating the need for CT simulation and error-prone image registration, ultimately reducing patient radiation dose and setup uncertainty. We propose an MRI-to-CT transformer-based denoising diffusion probabilistic model (MC-DDPM) to transform MRI into high-quality sCT to facilitate radiation treatment planning. MC-DDPM implements diffusion processes with a shifted-window transformer network to generate sCT from MRI. The proposed model consists of two processes: a forward process which adds Gaussian noise to real CT scans, and a reverse process in which a shifted-window transformer V-net (Swin-Vnet) denoises the noisy CT scans conditioned on the MRI from the same patient to produce noise-free CT scans. With an optimally trained Swin-Vnet, the reverse diffusion process was used to generate sCT scans matching MRI anatomy. We evaluated the proposed method by generating sCT from MRI on a brain dataset and a prostate dataset. Qualitative evaluation was performed using the mean absolute error (MAE) of Hounsfield unit (HU), peak signal to noise ratio (PSNR), multi-scale Structure Similarity index (MS-SSIM) and normalized cross correlation (NCC) indexes between ground truth CTs and sCTs. MC-DDPM generated brain sCTs with state-of-the-art quantitative results with MAE 43.317 HU, PSNR 27.046 dB, SSIM 0.965, and NCC 0.983. For the prostate dataset, MC-DDPM achieved MAE 59.953 HU, PSNR 26.920 dB, SSIM 0.849, and NCC 0.948. In conclusion, we have developed and validated a novel approach for generating CT images from routine MRIs using a transformer-based DDPM. This model effectively captures the complex relationship between CT and MRI images, allowing for robust and high-quality synthetic CT (sCT) images to be generated in minutes.
Cross-Shaped Windows Transformer with Self-supervised Pretraining for Clinically Significant Prostate Cancer Detection in Bi-parametric MRI
Apr 30, 2023



Abstract:Multiparametric magnetic resonance imaging (mpMRI) has demonstrated promising results in prostate cancer (PCa) detection using deep convolutional neural networks (CNNs). Recently, transformers have achieved competitive performance compared to CNNs in computer vision. Large-scale transformers need abundant annotated data for training, which are difficult to obtain in medical imaging. Self-supervised learning can effectively leverage unlabeled data to extract useful semantic representations without annotation and its associated costs. This can improve model performance on downstream tasks with limited labelled data and increase generalizability. We introduce a novel end-to-end Cross-Shaped windows (CSwin) transformer UNet model, CSwin UNet, to detect clinically significant prostate cancer (csPCa) in prostate bi-parametric MR imaging (bpMRI) and demonstrate the effectiveness of our proposed self-supervised pre-training framework. Using a large prostate bpMRI dataset with 1500 patients, we first pre-train CSwin transformer using multi-task self-supervised learning to improve data-efficiency and network generalizability. We then finetuned using lesion annotations to perform csPCa detection. Five-fold cross validation shows that self-supervised CSwin UNet achieves 0.888 AUC and 0.545 Average Precision (AP), significantly outperforming four state-of-the-art models (Swin UNETR, DynUNet, Attention UNet, UNet). Using a separate bpMRI dataset with 158 patients, we evaluated our model robustness to external hold-out data. Self-supervised CSwin UNet achieves 0.79 AUC and 0.45 AP, still outperforming all other comparable methods and demonstrating generalization to a dataset shift.
Cycle-guided Denoising Diffusion Probability Model for 3D Cross-modality MRI Synthesis
Apr 28, 2023Abstract:This study aims to develop a novel Cycle-guided Denoising Diffusion Probability Model (CG-DDPM) for cross-modality MRI synthesis. The CG-DDPM deploys two DDPMs that condition each other to generate synthetic images from two different MRI pulse sequences. The two DDPMs exchange random latent noise in the reverse processes, which helps to regularize both DDPMs and generate matching images in two modalities. This improves image-to-image translation ac-curacy. We evaluated the CG-DDPM quantitatively using mean absolute error (MAE), multi-scale structural similarity index measure (MSSIM), and peak sig-nal-to-noise ratio (PSNR), as well as the network synthesis consistency, on the BraTS2020 dataset. Our proposed method showed high accuracy and reliable consistency for MRI synthesis. In addition, we compared the CG-DDPM with several other state-of-the-art networks and demonstrated statistically significant improvements in the image quality of synthetic MRIs. The proposed method enhances the capability of current multimodal MRI synthesis approaches, which could contribute to more accurate diagnosis and better treatment planning for patients by synthesizing additional MRI modalities.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge