Mojtaba Safari
Efficient Vision Mamba for MRI Super-Resolution via Hybrid Selective Scanning
Dec 22, 2025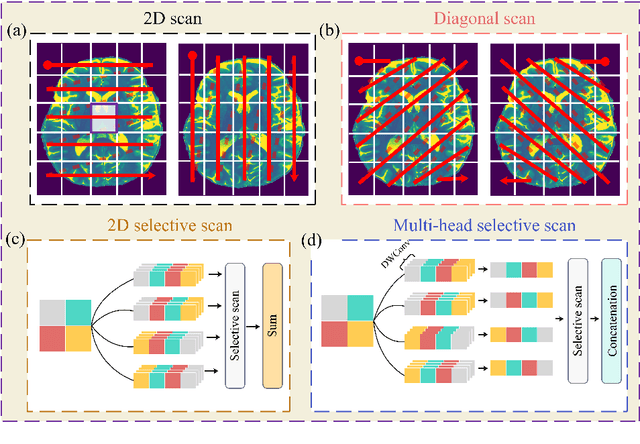
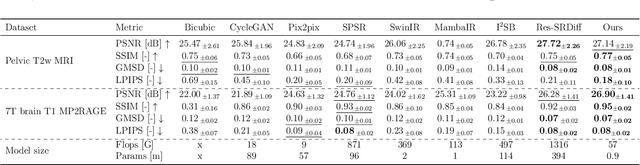
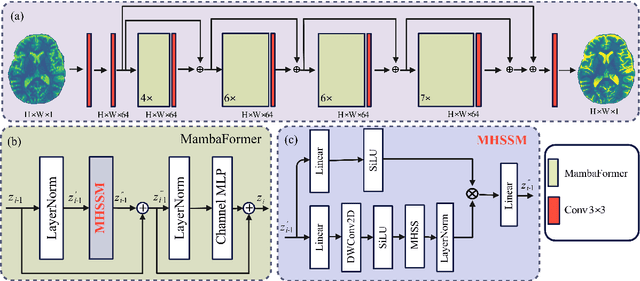

Abstract:Background: High-resolution MRI is critical for diagnosis, but long acquisition times limit clinical use. Super-resolution (SR) can enhance resolution post-scan, yet existing deep learning methods face fidelity-efficiency trade-offs. Purpose: To develop a computationally efficient and accurate deep learning framework for MRI SR that preserves anatomical detail for clinical integration. Materials and Methods: We propose a novel SR framework combining multi-head selective state-space models (MHSSM) with a lightweight channel MLP. The model uses 2D patch extraction with hybrid scanning to capture long-range dependencies. Each MambaFormer block integrates MHSSM, depthwise convolutions, and gated channel mixing. Evaluation used 7T brain T1 MP2RAGE maps (n=142) and 1.5T prostate T2w MRI (n=334). Comparisons included Bicubic interpolation, GANs (CycleGAN, Pix2pix, SPSR), transformers (SwinIR), Mamba (MambaIR), and diffusion models (I2SB, Res-SRDiff). Results: Our model achieved superior performance with exceptional efficiency. For 7T brain data: SSIM=0.951+-0.021, PSNR=26.90+-1.41 dB, LPIPS=0.076+-0.022, GMSD=0.083+-0.017, significantly outperforming all baselines (p<0.001). For prostate data: SSIM=0.770+-0.049, PSNR=27.15+-2.19 dB, LPIPS=0.190+-0.095, GMSD=0.087+-0.013. The framework used only 0.9M parameters and 57 GFLOPs, reducing parameters by 99.8% and computation by 97.5% versus Res-SRDiff, while outperforming SwinIR and MambaIR in accuracy and efficiency. Conclusion: The proposed framework provides an efficient, accurate MRI SR solution, delivering enhanced anatomical detail across datasets. Its low computational demand and state-of-the-art performance show strong potential for clinical translation.
Explainable Cross-Disease Reasoning for Cardiovascular Risk Assessment from LDCT
Nov 13, 2025Abstract:Low-dose chest computed tomography (LDCT) inherently captures both pulmonary and cardiac structures, offering a unique opportunity for joint assessment of lung and cardiovascular health. However, most existing approaches treat these domains as independent tasks, overlooking their physiological interplay and shared imaging biomarkers. We propose an Explainable Cross-Disease Reasoning Framework that enables interpretable cardiopulmonary risk assessment from a single LDCT scan. The framework introduces an agentic reasoning process that emulates clinical diagnostic thinking-first perceiving pulmonary findings, then reasoning through established medical knowledge, and finally deriving a cardiovascular judgment with explanatory rationale. It integrates three synergistic components: a pulmonary perception module that summarizes lung abnormalities, a knowledge-guided reasoning module that infers their cardiovascular implications, and a cardiac representation module that encodes structural biomarkers. Their outputs are fused to produce a holistic cardiovascular risk prediction that is both accurate and physiologically grounded. Experiments on the NLST cohort demonstrate that the proposed framework achieves state-of-the-art performance for CVD screening and mortality prediction, outperforming single-disease and purely image-based baselines. Beyond quantitative gains, the framework provides human-verifiable reasoning that aligns with cardiological understanding, revealing coherent links between pulmonary abnormalities and cardiac stress mechanisms. Overall, this work establishes a unified and explainable paradigm for cardiovascular analysis from LDCT, bridging the gap between image-based prediction and mechanism-based medical interpretation.
Systematic Review and Meta-analysis of AI-driven MRI Motion Artifact Detection and Correction
Sep 05, 2025Abstract:Background: To systematically review and perform a meta-analysis of artificial intelligence (AI)-driven methods for detecting and correcting magnetic resonance imaging (MRI) motion artifacts, assessing current developments, effectiveness, challenges, and future research directions. Methods: A comprehensive systematic review and meta-analysis were conducted, focusing on deep learning (DL) approaches, particularly generative models, for the detection and correction of MRI motion artifacts. Quantitative data were extracted regarding utilized datasets, DL architectures, and performance metrics. Results: DL, particularly generative models, show promise for reducing motion artifacts and improving image quality; however, limited generalizability, reliance on paired training data, and risk of visual distortions remain key challenges that motivate standardized datasets and reporting. Conclusions: AI-driven methods, particularly DL generative models, show significant potential for improving MRI image quality by effectively addressing motion artifacts. However, critical challenges must be addressed, including the need for comprehensive public datasets, standardized reporting protocols for artifact levels, and more advanced, adaptable DL techniques to reduce reliance on extensive paired datasets. Addressing these aspects could substantially enhance MRI diagnostic accuracy, reduce healthcare costs, and improve patient care outcomes.
DINOv3 with Test-Time Training for Medical Image Registration
Aug 20, 2025Abstract:Prior medical image registration approaches, particularly learning-based methods, often require large amounts of training data, which constrains clinical adoption. To overcome this limitation, we propose a training-free pipeline that relies on a frozen DINOv3 encoder and test-time optimization of the deformation field in feature space. Across two representative benchmarks, the method is accurate and yields regular deformations. On Abdomen MR-CT, it attained the best mean Dice score (DSC) of 0.790 together with the lowest 95th percentile Hausdorff Distance (HD95) of 4.9+-5.0 and the lowest standard deviation of Log-Jacobian (SDLogJ) of 0.08+-0.02. On ACDC cardiac MRI, it improves mean DSC to 0.769 and reduces SDLogJ to 0.11 and HD95 to 4.8, a marked gain over the initial alignment. The results indicate that operating in a compact foundation feature space at test time offers a practical and general solution for clinical registration without additional training.
Capabilities of GPT-5 on Multimodal Medical Reasoning
Aug 13, 2025Abstract:Recent advances in large language models (LLMs) have enabled general-purpose systems to perform increasingly complex domain-specific reasoning without extensive fine-tuning. In the medical domain, decision-making often requires integrating heterogeneous information sources, including patient narratives, structured data, and medical images. This study positions GPT-5 as a generalist multimodal reasoner for medical decision support and systematically evaluates its zero-shot chain-of-thought reasoning performance on both text-based question answering and visual question answering tasks under a unified protocol. We benchmark GPT-5, GPT-5-mini, GPT-5-nano, and GPT-4o-2024-11-20 against standardized splits of MedQA, MedXpertQA (text and multimodal), MMLU medical subsets, USMLE self-assessment exams, and VQA-RAD. Results show that GPT-5 consistently outperforms all baselines, achieving state-of-the-art accuracy across all QA benchmarks and delivering substantial gains in multimodal reasoning. On MedXpertQA MM, GPT-5 improves reasoning and understanding scores by +29.26% and +26.18% over GPT-4o, respectively, and surpasses pre-licensed human experts by +24.23% in reasoning and +29.40% in understanding. In contrast, GPT-4o remains below human expert performance in most dimensions. A representative case study demonstrates GPT-5's ability to integrate visual and textual cues into a coherent diagnostic reasoning chain, recommending appropriate high-stakes interventions. Our results show that, on these controlled multimodal reasoning benchmarks, GPT-5 moves from human-comparable to above human-expert performance. This improvement may substantially inform the design of future clinical decision-support systems.
MRI motion correction via efficient residual-guided denoising diffusion probabilistic models
May 06, 2025Abstract:Purpose: Motion artifacts in magnetic resonance imaging (MRI) significantly degrade image quality and impair quantitative analysis. Conventional mitigation strategies, such as repeated acquisitions or motion tracking, are costly and workflow-intensive. This study introduces Res-MoCoDiff, an efficient denoising diffusion probabilistic model tailored for MRI motion artifact correction. Methods: Res-MoCoDiff incorporates a novel residual error shifting mechanism in the forward diffusion process, aligning the noise distribution with motion-corrupted data and enabling an efficient four-step reverse diffusion. A U-net backbone enhanced with Swin-Transformer blocks conventional attention layers, improving adaptability across resolutions. Training employs a combined l1+l2 loss, which promotes image sharpness and reduces pixel-level errors. Res-MoCoDiff was evaluated on synthetic dataset generated using a realistic motion simulation framework and on an in-vivo dataset. Comparative analyses were conducted against established methods, including CycleGAN, Pix2pix, and MT-DDPM using quantitative metrics such as peak signal-to-noise ratio (PSNR), structural similarity index measure (SSIM), and normalized mean squared error (NMSE). Results: The proposed method demonstrated superior performance in removing motion artifacts across all motion severity levels. Res-MoCoDiff consistently achieved the highest SSIM and the lowest NMSE values, with a PSNR of up to 41.91+-2.94 dB for minor distortions. Notably, the average sampling time was reduced to 0.37 seconds per batch of two image slices, compared with 101.74 seconds for conventional approaches.
MRI super-resolution reconstruction using efficient diffusion probabilistic model with residual shifting
Mar 03, 2025Abstract:Objective:This study introduces a residual error-shifting mechanism that drastically reduces sampling steps while preserving critical anatomical details, thus accelerating MRI reconstruction. Approach:We propose a novel diffusion-based SR framework called Res-SRDiff, which integrates residual error shifting into the forward diffusion process. This enables efficient HR image reconstruction by aligning the degraded HR and LR distributions.We evaluated Res-SRDiff on ultra-high-field brain T1 MP2RAGE maps and T2-weighted prostate images, comparing it with Bicubic, Pix2pix, CycleGAN, and a conventional denoising diffusion probabilistic model with vision transformer backbone (TM-DDPM), using quantitative metrics such as peak signal-to-noise ratio (PSNR), structural similarity index (SSIM), gradient magnitude similarity deviation (GMSD), and learned perceptual image patch similarity (LPIPS). Main results: Res-SRDiff significantly outperformed all comparative methods in terms of PSNR, SSIM, and GMSD across both datasets, with statistically significant improvements (p-values<<0.05). The model achieved high-fidelity image restoration with only four sampling steps, drastically reducing computational time to under one second per slice, which is substantially faster than conventional TM-DDPM with around 20 seconds per slice. Qualitative analyses further demonstrated that Res-SRDiff effectively preserved fine anatomical details and lesion morphology in both brain and pelvic MRI images. Significance: Our findings show that Res-SRDiff is an efficient and accurate MRI SR method, markedly improving computational efficiency and image quality. Integrating residual error shifting into the diffusion process allows for rapid and robust HR image reconstruction, enhancing clinical MRI workflows and advancing medical imaging research. The source at:https://github.com/mosaf/Res-SRDiff
Triad: Vision Foundation Model for 3D Magnetic Resonance Imaging
Feb 23, 2025Abstract:Vision foundation models (VFMs) are pre-trained on extensive image datasets to learn general representations for diverse types of data. These models can subsequently be fine-tuned for specific downstream tasks, significantly boosting performance across a broad range of applications. However, existing vision foundation models that claim to be applicable to various clinical tasks are mostly pre-trained on 3D computed tomography (CT), which benefits from the availability of extensive 3D CT databases. Significant differences between CT and magnetic resonance imaging (MRI) in imaging principles, signal characteristics, and data distribution may hinder their practical performance and versatility in MRI-specific applications. Here, we propose Triad, a vision foundation model for 3D MRI. Triad adopts a widely used autoencoder architecture to learn robust representations from 131,170 3D MRI volumes and uses organ-independent imaging descriptions to constrain the semantic distribution of the visual modality. The above pre-training dataset is called Triad-131K, which is currently the largest 3D MRI pre-training dataset. We evaluate Triad across three tasks, namely, organ/tumor segmentation, organ/cancer classification, and medical image registration, in two data modalities (within-domain and out-of-domain) settings using 25 downstream datasets. By initializing models with Triad's pre-trained weights, nnUNet-Triad improves segmentation performance by 2.51% compared to nnUNet-Scratch across 17 datasets. Swin-B-Triad achieves a 3.97% improvement over Swin-B-Scratch in classification tasks across five datasets. SwinUNETR-Triad improves by 4.00% compared to SwinUNETR-Scratch in registration tasks across two datasets. Our study demonstrates that pre-training can improve performance when the data modalities and organs of upstream and downstream tasks are consistent.
A Physics-Informed Deep Learning Model for MRI Brain Motion Correction
Feb 13, 2025Abstract:Background: MRI is crucial for brain imaging but is highly susceptible to motion artifacts due to long acquisition times. This study introduces PI-MoCoNet, a physics-informed motion correction network that integrates spatial and k-space information to remove motion artifacts without explicit motion parameter estimation, enhancing image fidelity and diagnostic reliability. Materials and Methods: PI-MoCoNet consists of a motion detection network (U-net with spatial averaging) to identify corrupted k-space lines and a motion correction network (U-net with Swin Transformer blocks) to reconstruct motion-free images. The correction is guided by three loss functions: reconstruction (L1), perceptual (LPIPS), and data consistency (Ldc). Motion artifacts were simulated via rigid phase encoding perturbations and evaluated on IXI and MR-ART datasets against Pix2Pix, CycleGAN, and U-net using PSNR, SSIM, and NMSE. Results: PI-MoCoNet significantly improved image quality. On IXI, for minor artifacts, PSNR increased from 34.15 dB to 45.95 dB, SSIM from 0.87 to 1.00, and NMSE reduced from 0.55% to 0.04%. For moderate artifacts, PSNR improved from 30.23 dB to 42.16 dB, SSIM from 0.80 to 0.99, and NMSE from 1.32% to 0.09%. For heavy artifacts, PSNR rose from 27.99 dB to 36.01 dB, SSIM from 0.75 to 0.97, and NMSE decreased from 2.21% to 0.36%. On MR-ART, PI-MoCoNet achieved PSNR gains of ~10 dB and SSIM improvements of up to 0.20, with NMSE reductions of ~6%. Ablation studies confirmed the importance of data consistency and perceptual losses, yielding a 1 dB PSNR gain and 0.17% NMSE reduction. Conclusions: PI-MoCoNet effectively mitigates motion artifacts in brain MRI, outperforming existing methods. Its ability to integrate spatial and k-space information makes it a promising tool for clinical use in motion-prone settings. Code: https://github.com/mosaf/PI-MoCoNet.git.
Advancing MRI Reconstruction: A Systematic Review of Deep Learning and Compressed Sensing Integration
Jan 24, 2025
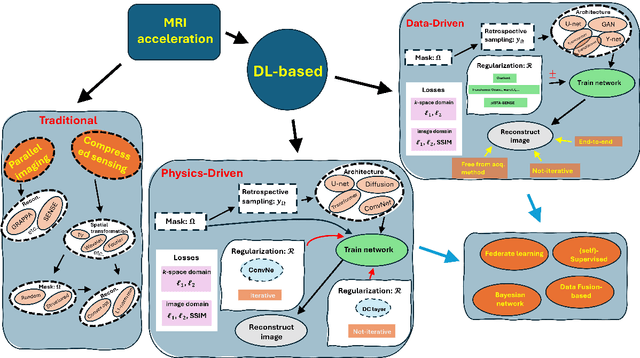
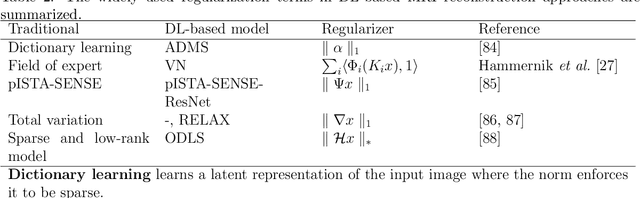
Abstract:Magnetic resonance imaging (MRI) is a non-invasive imaging modality and provides comprehensive anatomical and functional insights into the human body. However, its long acquisition times can lead to patient discomfort, motion artifacts, and limiting real-time applications. To address these challenges, strategies such as parallel imaging have been applied, which utilize multiple receiver coils to speed up the data acquisition process. Additionally, compressed sensing (CS) is a method that facilitates image reconstruction from sparse data, significantly reducing image acquisition time by minimizing the amount of data collection needed. Recently, deep learning (DL) has emerged as a powerful tool for improving MRI reconstruction. It has been integrated with parallel imaging and CS principles to achieve faster and more accurate MRI reconstructions. This review comprehensively examines DL-based techniques for MRI reconstruction. We categorize and discuss various DL-based methods, including end-to-end approaches, unrolled optimization, and federated learning, highlighting their potential benefits. Our systematic review highlights significant contributions and underscores the potential of DL in MRI reconstruction. Additionally, we summarize key results and trends in DL-based MRI reconstruction, including quantitative metrics, the dataset, acceleration factors, and the progress of and research interest in DL techniques over time. Finally, we discuss potential future directions and the importance of DL-based MRI reconstruction in advancing medical imaging. To facilitate further research in this area, we provide a GitHub repository that includes up-to-date DL-based MRI reconstruction publications and public datasets-https://github.com/mosaf/Awesome-DL-based-CS-MRI.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge