Richard Baumgartner
Challenges in Variable Importance Ranking Under Correlation
Feb 05, 2024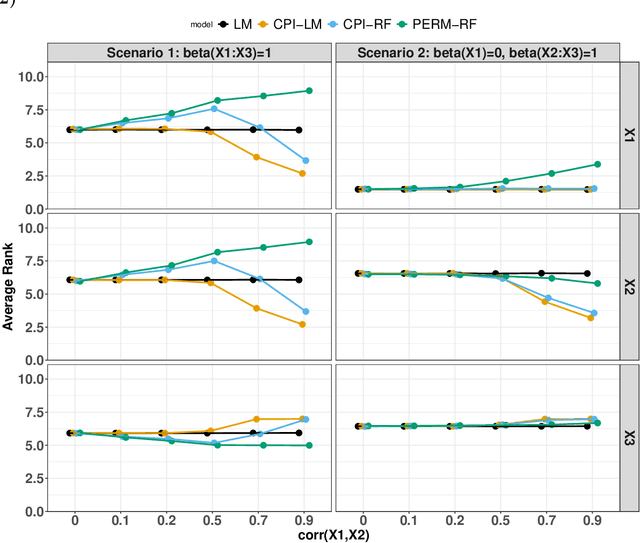
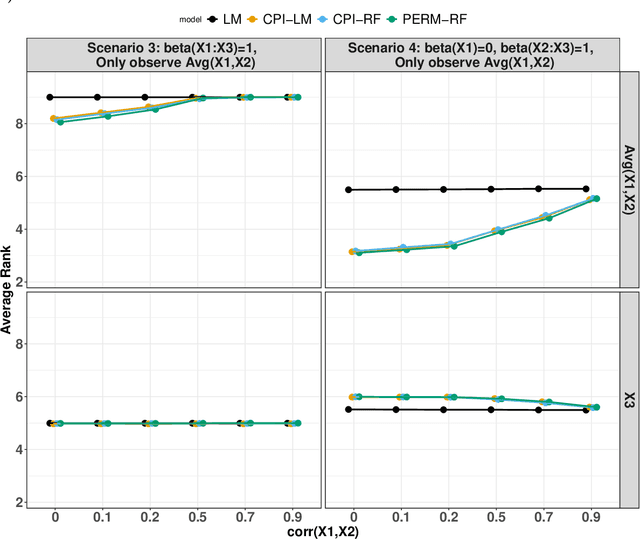
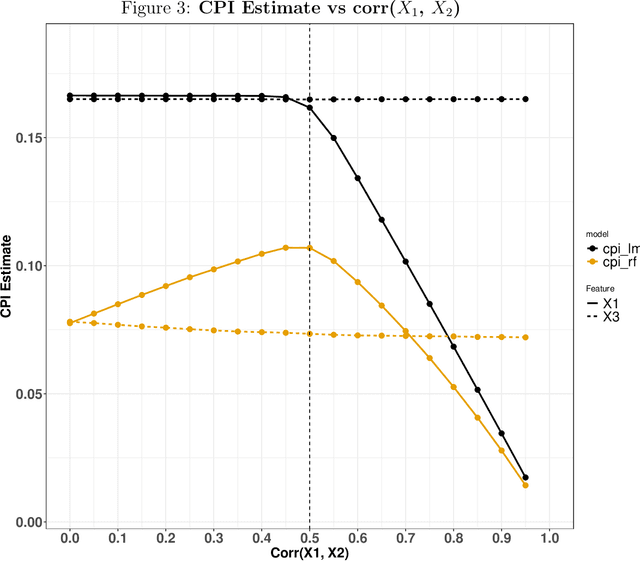
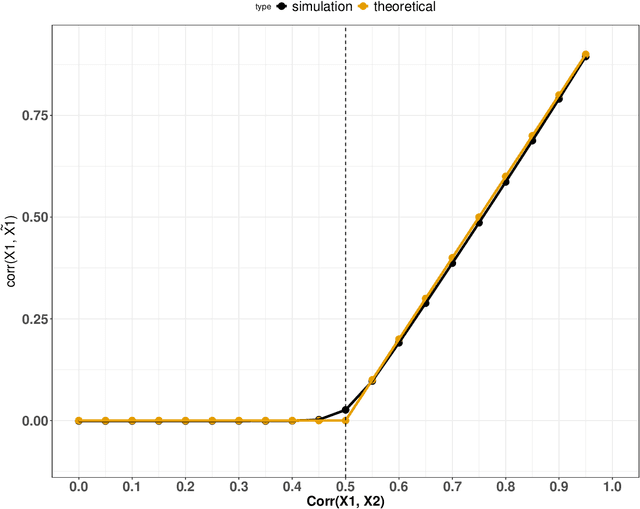
Abstract:Variable importance plays a pivotal role in interpretable machine learning as it helps measure the impact of factors on the output of the prediction model. Model agnostic methods based on the generation of "null" features via permutation (or related approaches) can be applied. Such analysis is often utilized in pharmaceutical applications due to its ability to interpret black-box models, including tree-based ensembles. A major challenge and significant confounder in variable importance estimation however is the presence of between-feature correlation. Recently, several adjustments to marginal permutation utilizing feature knockoffs were proposed to address this issue, such as the variable importance measure known as conditional predictive impact (CPI). Assessment and evaluation of such approaches is the focus of our work. We first present a comprehensive simulation study investigating the impact of feature correlation on the assessment of variable importance. We then theoretically prove the limitation that highly correlated features pose for the CPI through the knockoff construction. While we expect that there is always no correlation between knockoff variables and its corresponding predictor variables, we prove that the correlation increases linearly beyond a certain correlation threshold between the predictor variables. Our findings emphasize the absence of free lunch when dealing with high feature correlation, as well as the necessity of understanding the utility and limitations behind methods in variable importance estimation.
Multi-dimension unified Swin Transformer for 3D Lesion Segmentation in Multiple Anatomical Locations
Sep 04, 2023Abstract:In oncology research, accurate 3D segmentation of lesions from CT scans is essential for the modeling of lesion growth kinetics. However, following the RECIST criteria, radiologists routinely only delineate each lesion on the axial slice showing the largest transverse area, and delineate a small number of lesions in 3D for research purposes. As a result, we have plenty of unlabeled 3D volumes and labeled 2D images, and scarce labeled 3D volumes, which makes training a deep-learning 3D segmentation model a challenging task. In this work, we propose a novel model, denoted a multi-dimension unified Swin transformer (MDU-ST), for 3D lesion segmentation. The MDU-ST consists of a Shifted-window transformer (Swin-transformer) encoder and a convolutional neural network (CNN) decoder, allowing it to adapt to 2D and 3D inputs and learn the corresponding semantic information in the same encoder. Based on this model, we introduce a three-stage framework: 1) leveraging large amount of unlabeled 3D lesion volumes through self-supervised pretext tasks to learn the underlying pattern of lesion anatomy in the Swin-transformer encoder; 2) fine-tune the Swin-transformer encoder to perform 2D lesion segmentation with 2D RECIST slices to learn slice-level segmentation information; 3) further fine-tune the Swin-transformer encoder to perform 3D lesion segmentation with labeled 3D volumes. The network's performance is evaluated by the Dice similarity coefficient (DSC) and Hausdorff distance (HD) using an internal 3D lesion dataset with 593 lesions extracted from multiple anatomical locations. The proposed MDU-ST demonstrates significant improvement over the competing models. The proposed method can be used to conduct automated 3D lesion segmentation to assist radiomics and tumor growth modeling studies. This paper has been accepted by the IEEE International Symposium on Biomedical Imaging (ISBI) 2023.
Target alignment in truncated kernel ridge regression
Jun 28, 2022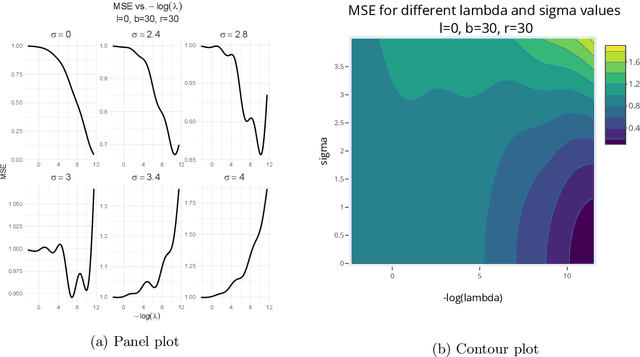
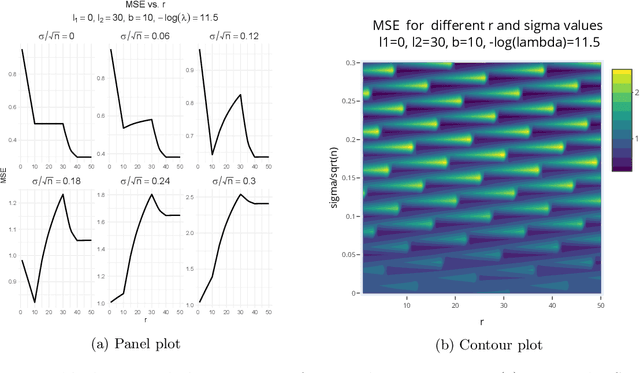
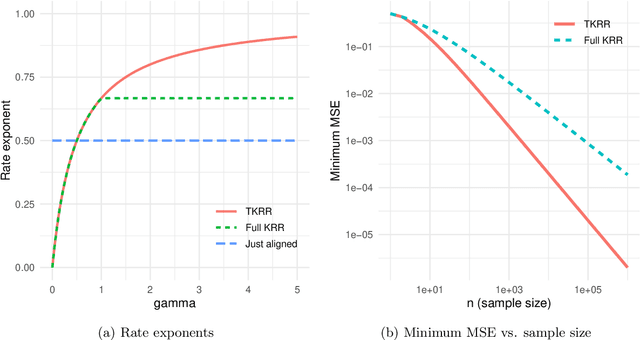
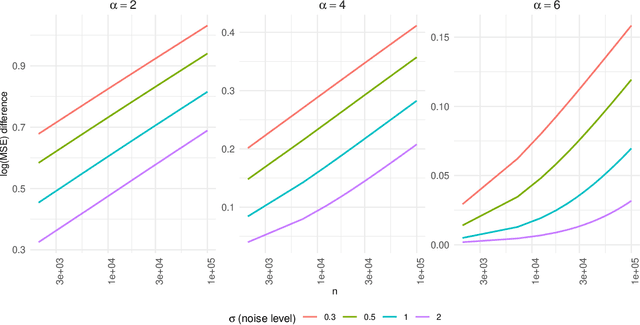
Abstract:Kernel ridge regression (KRR) has recently attracted renewed interest due to its potential for explaining the transient effects, such as double descent, that emerge during neural network training. In this work, we study how the alignment between the target function and the kernel affects the performance of the KRR. We focus on the truncated KRR (TKRR) which utilizes an additional parameter that controls the spectral truncation of the kernel matrix. We show that for polynomial alignment, there is an \emph{over-aligned} regime, in which TKRR can achieve a faster rate than what is achievable by full KRR. The rate of TKRR can improve all the way to the parametric rate, while that of full KRR is capped at a sub-optimal value. This shows that target alignemnt can be better leveraged by utilizing spectral truncation in kernel methods. We also consider the bandlimited alignment setting and show that the regularization surface of TKRR can exhibit transient effects including multiple descent and non-monotonic behavior. Our results show that there is a strong and quantifable relation between the shape of the \emph{alignment spectrum} and the generalization performance of kernel methods, both in terms of rates and in finite samples.
A Framework for an Assessment of the Kernel-target Alignment in Tree Ensemble Kernel Learning
Aug 19, 2021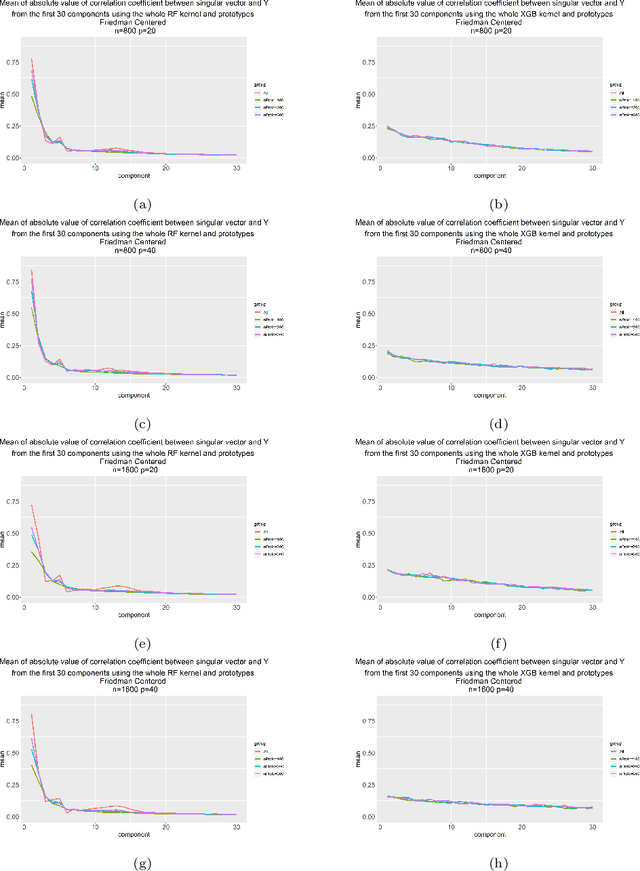
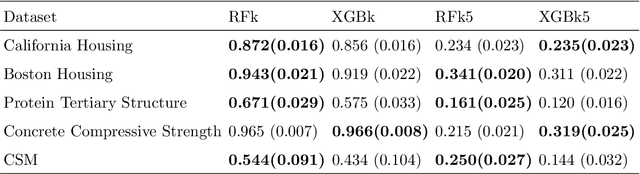
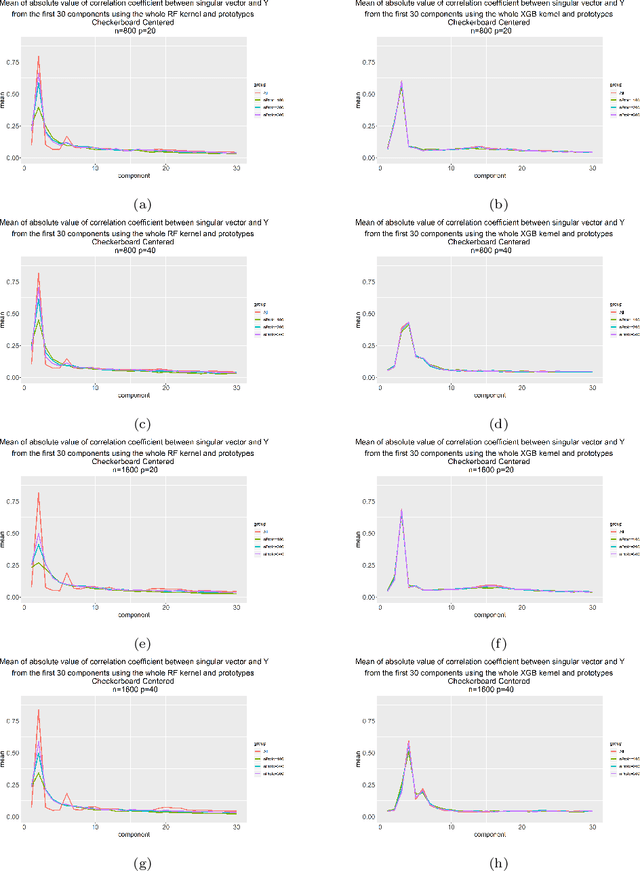
Abstract:Kernels ensuing from tree ensembles such as random forest (RF) or gradient boosted trees (GBT), when used for kernel learning, have been shown to be competitive to their respective tree ensembles (particularly in higher dimensional scenarios). On the other hand, it has been also shown that performance of the kernel algorithms depends on the degree of the kernel-target alignment. However, the kernel-target alignment for kernel learning based on the tree ensembles has not been investigated and filling this gap is the main goal of our work. Using the eigenanalysis of the kernel matrix, we demonstrate that for continuous targets good performance of the tree-based kernel learning is associated with strong kernel-target alignment. Moreover, we show that well performing tree ensemble based kernels are characterized by strong target aligned components that are expressed through scalar products between the eigenvectors of the kernel matrix and the target. This suggests that when tree ensemble based kernel learning is successful, relevant information for the supervised problem is concentrated near lower dimensional manifold spanned by the target aligned components. Persistence of the strong target aligned components in tree ensemble based kernels is further supported by sensitivity analysis via landmark learning. In addition to a comprehensive simulation study, we also provide experimental results from several real life data sets that are in line with the simulations.
(Decision and regression) tree ensemble based kernels for regression and classification
Dec 19, 2020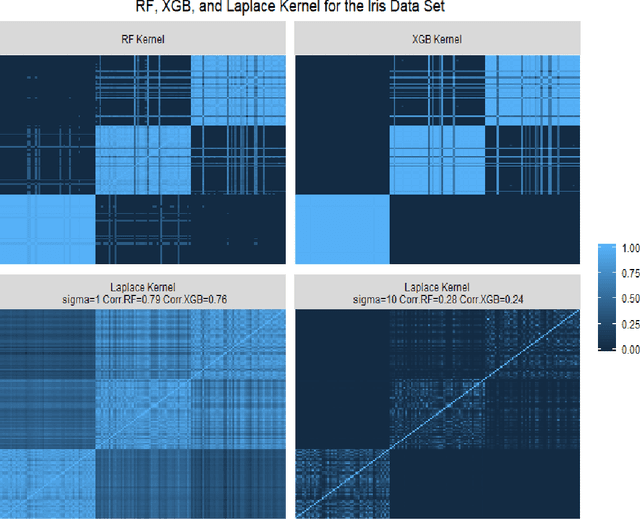
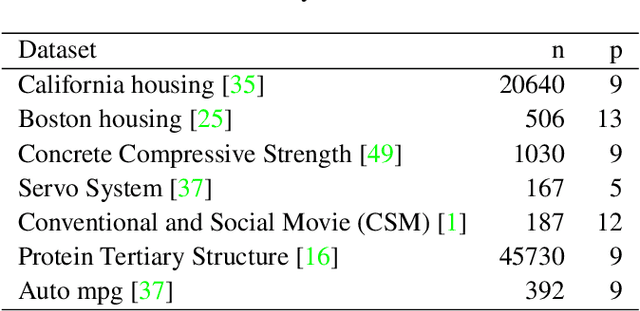
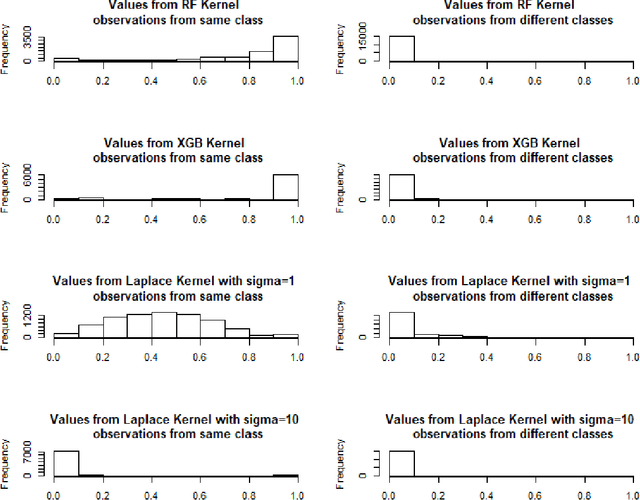
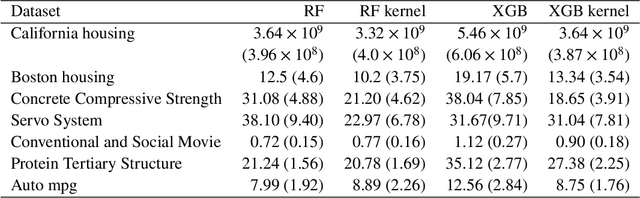
Abstract:Tree based ensembles such as Breiman's random forest (RF) and Gradient Boosted Trees (GBT) can be interpreted as implicit kernel generators, where the ensuing proximity matrix represents the data-driven tree ensemble kernel. Kernel perspective on the RF has been used to develop a principled framework for theoretical investigation of its statistical properties. Recently, it has been shown that the kernel interpretation is germane to other tree-based ensembles e.g. GBTs. However, practical utility of the links between kernels and the tree ensembles has not been widely explored and systematically evaluated. Focus of our work is investigation of the interplay between kernel methods and the tree based ensembles including the RF and GBT. We elucidate the performance and properties of the RF and GBT based kernels in a comprehensive simulation study comprising of continuous and binary targets. We show that for continuous targets, the RF/GBT kernels are competitive to their respective ensembles in higher dimensional scenarios, particularly in cases with larger number of noisy features. For the binary target, the RF/GBT kernels and their respective ensembles exhibit comparable performance. We provide the results from real life data sets for regression and classification to show how these insights may be leveraged in practice. Overall, our results support the tree ensemble based kernels as a valuable addition to the practitioner's toolbox. Finally, we discuss extensions of the tree ensemble based kernels for survival targets, interpretable prototype and landmarking classification and regression. We outline future line of research for kernels furnished by Bayesian counterparts of the frequentist tree ensembles.
Random Forest (RF) Kernel for Regression, Classification and Survival
Aug 31, 2020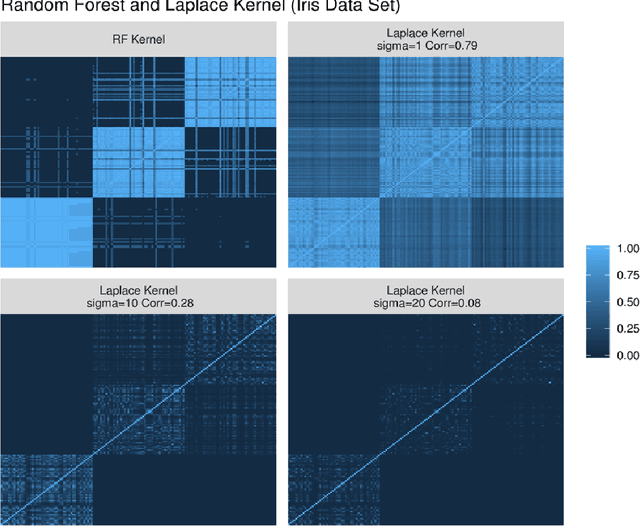
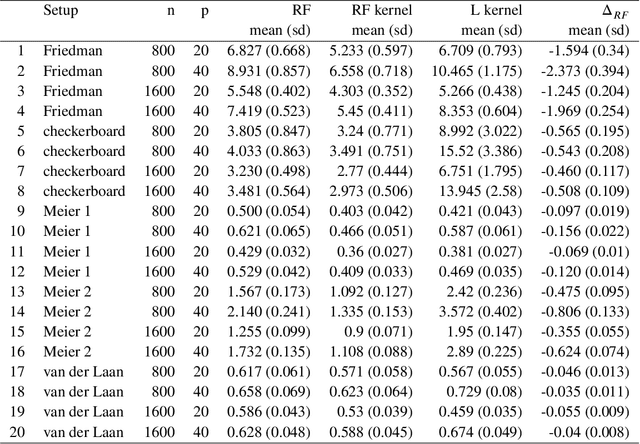
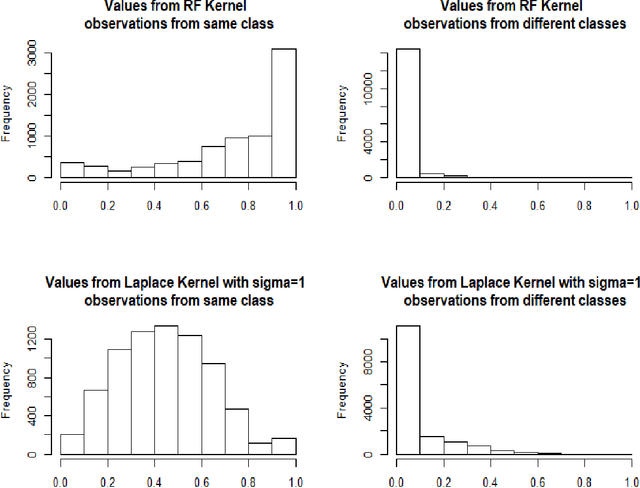
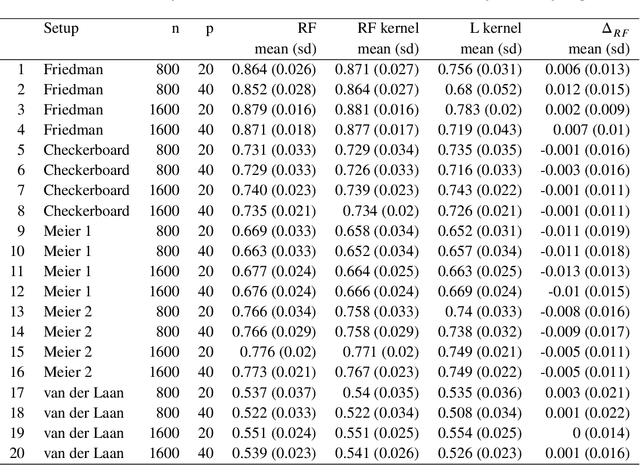
Abstract:Breiman's random forest (RF) can be interpreted as an implicit kernel generator,where the ensuing proximity matrix represents the data-driven RF kernel. Kernel perspective on the RF has been used to develop a principled framework for theoretical investigation of its statistical properties. However, practical utility of the links between kernels and the RF has not been widely explored and systematically evaluated.Focus of our work is investigation of the interplay between kernel methods and the RF. We elucidate the performance and properties of the data driven RF kernels used by regularized linear models in a comprehensive simulation study comprising of continuous, binary and survival targets. We show that for continuous and survival targets, the RF kernels are competitive to RF in higher dimensional scenarios with larger number of noisy features. For the binary target, the RF kernel and RF exhibit comparable performance. As the RF kernel asymptotically converges to the Laplace kernel, we included it in our evaluation. For most simulation setups, the RF and RFkernel outperformed the Laplace kernel. Nevertheless, in some cases the Laplace kernel was competitive, showing its potential value for applications. We also provide the results from real life data sets for the regression, classification and survival to illustrate how these insights may be leveraged in practice.Finally, we discuss further extensions of the RF kernels in the context of interpretable prototype and landmarking classification, regression and survival. We outline future line of research for kernels furnished by Bayesian counterparts of the RF.
A deep learning-facilitated radiomics solution for the prediction of lung lesion shrinkage in non-small cell lung cancer trials
Mar 05, 2020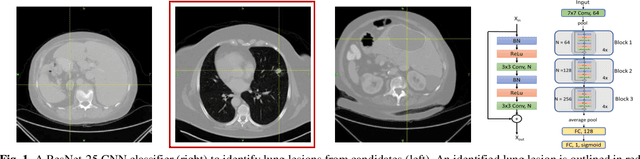
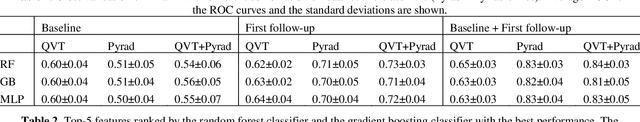
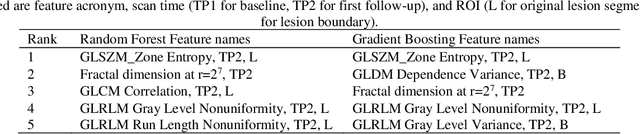
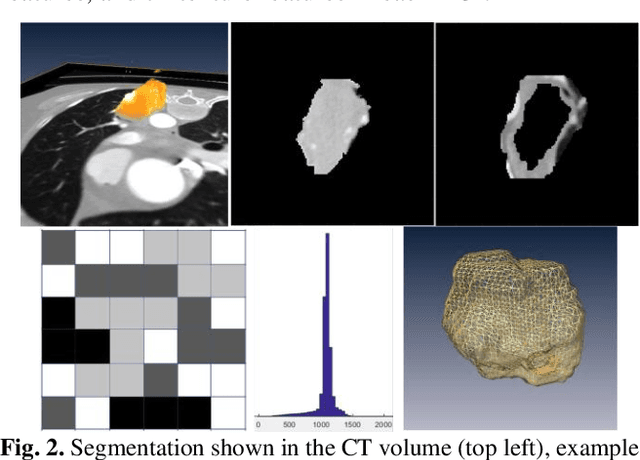
Abstract:Herein we propose a deep learning-based approach for the prediction of lung lesion response based on radiomic features extracted from clinical CT scans of patients in non-small cell lung cancer trials. The approach starts with the classification of lung lesions from the set of primary and metastatic lesions at various anatomic locations. Focusing on the lung lesions, we perform automatic segmentation to extract their 3D volumes. Radiomic features are then extracted from the lesion on the pre-treatment scan and the first follow-up scan to predict which lesions will shrink at least 30% in diameter during treatment (either Pembrolizumab or combinations of chemotherapy and Pembrolizumab), which is defined as a partial response by the Response Evaluation Criteria In Solid Tumors (RECIST) guidelines. A 5-fold cross validation on the training set led to an AUC of 0.84 +/- 0.03, and the prediction on the testing dataset reached AUC of 0.73 +/- 0.02 for the outcome of 30% diameter shrinkage.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge