Yabo Fu
Spatiotemporal Gaussian Optimization for 4D Cone Beam CT Reconstruction from Sparse Projections
Jan 07, 2025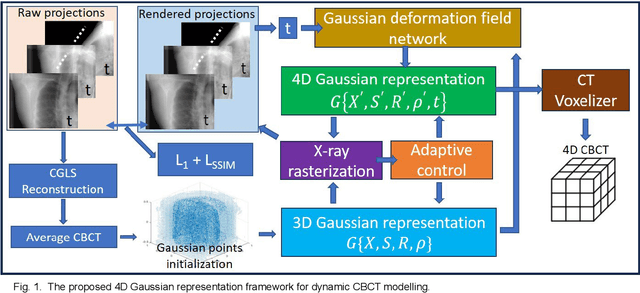
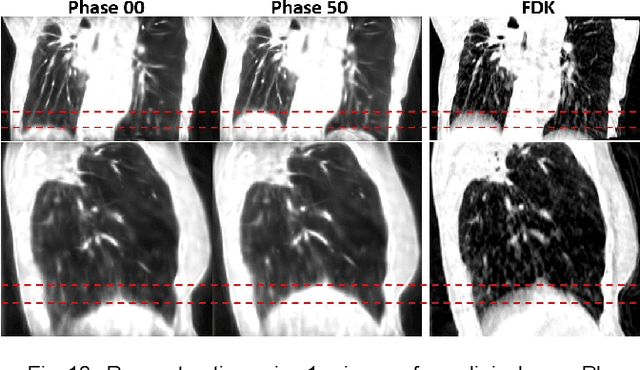
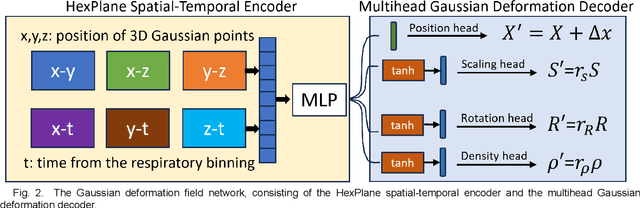
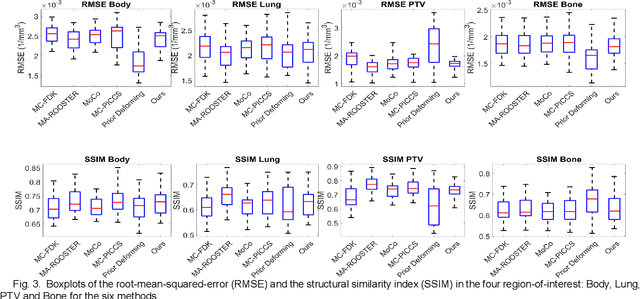
Abstract:In image-guided radiotherapy (IGRT), four-dimensional cone-beam computed tomography (4D-CBCT) is critical for assessing tumor motion during a patients breathing cycle prior to beam delivery. However, generating 4D-CBCT images with sufficient quality requires significantly more projection images than a standard 3D-CBCT scan, leading to extended scanning times and increased imaging dose to the patient. To address these limitations, there is a strong demand for methods capable of reconstructing high-quality 4D-CBCT images from a 1-minute 3D-CBCT acquisition. The challenge lies in the sparse sampling of projections, which introduces severe streaking artifacts and compromises image quality. This paper introduces a novel framework leveraging spatiotemporal Gaussian representation for 4D-CBCT reconstruction from sparse projections, achieving a balance between streak artifact reduction, dynamic motion preservation, and fine detail restoration. Each Gaussian is characterized by its 3D position, covariance, rotation, and density. Two-dimensional X-ray projection images can be rendered from the Gaussian point cloud representation via X-ray rasterization. The properties of each Gaussian were optimized by minimizing the discrepancy between the measured projections and the rendered X-ray projections. A Gaussian deformation network is jointly optimized to deform these Gaussian properties to obtain a 4D Gaussian representation for dynamic CBCT scene modeling. The final 4D-CBCT images are reconstructed by voxelizing the 4D Gaussians, achieving a high-quality representation that preserves both motion dynamics and spatial detail. The code and reconstruction results can be found at https://github.com/fuyabo/4DGS_for_4DCBCT/tree/main
Deformable Image Registration using Unsupervised Deep Learning for CBCT-guided Abdominal Radiotherapy
Aug 29, 2022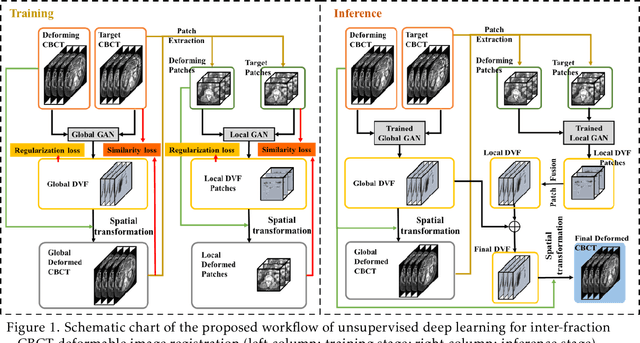
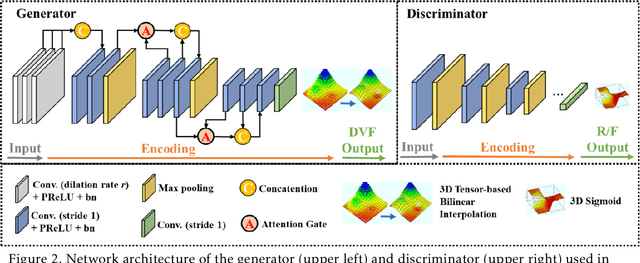
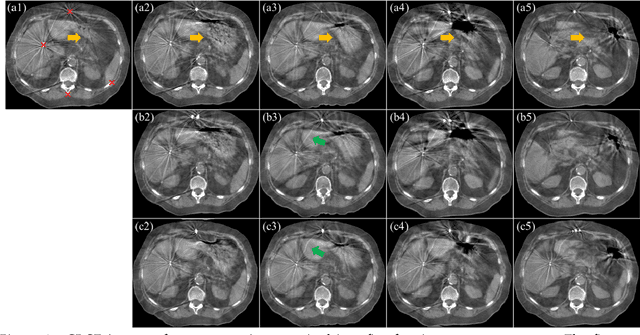
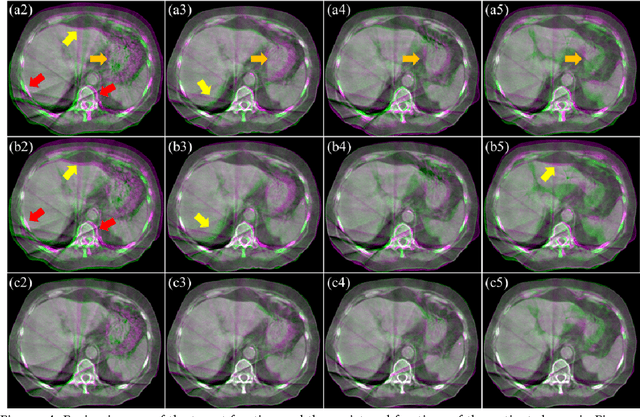
Abstract:CBCTs in image-guided radiotherapy provide crucial anatomy information for patient setup and plan evaluation. Longitudinal CBCT image registration could quantify the inter-fractional anatomic changes. The purpose of this study is to propose an unsupervised deep learning based CBCT-CBCT deformable image registration. The proposed deformable registration workflow consists of training and inference stages that share the same feed-forward path through a spatial transformation-based network (STN). The STN consists of a global generative adversarial network (GlobalGAN) and a local GAN (LocalGAN) to predict the coarse- and fine-scale motions, respectively. The network was trained by minimizing the image similarity loss and the deformable vector field (DVF) regularization loss without the supervision of ground truth DVFs. During the inference stage, patches of local DVF were predicted by the trained LocalGAN and fused to form a whole-image DVF. The local whole-image DVF was subsequently combined with the GlobalGAN generated DVF to obtain final DVF. The proposed method was evaluated using 100 fractional CBCTs from 20 abdominal cancer patients in the experiments and 105 fractional CBCTs from a cohort of 21 different abdominal cancer patients in a holdout test. Qualitatively, the registration results show great alignment between the deformed CBCT images and the target CBCT image. Quantitatively, the average target registration error (TRE) calculated on the fiducial markers and manually identified landmarks was 1.91+-1.11 mm. The average mean absolute error (MAE), normalized cross correlation (NCC) between the deformed CBCT and target CBCT were 33.42+-7.48 HU, 0.94+-0.04, respectively. This promising registration method could provide fast and accurate longitudinal CBCT alignment to facilitate inter-fractional anatomic changes analysis and prediction.
Deep Learning in Multi-organ Segmentation
Jan 28, 2020
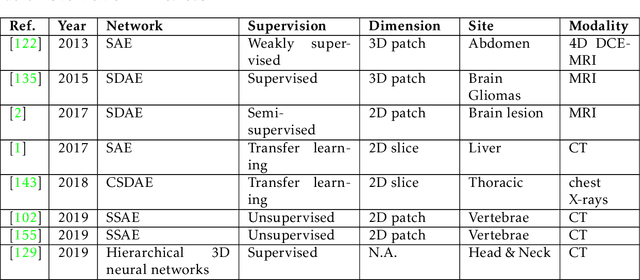
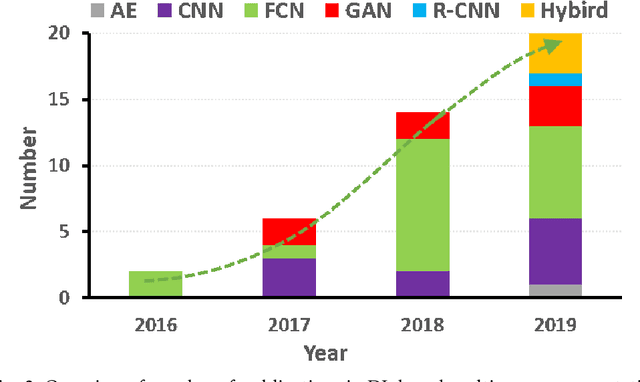

Abstract:This paper presents a review of deep learning (DL) in multi-organ segmentation. We summarized the latest DL-based methods for medical image segmentation and applications. These methods were classified into six categories according to their network design. For each category, we listed the surveyed works, highlighted important contributions and identified specific challenges. Following the detailed review of each category, we briefly discussed its achievements, shortcomings and future potentials. We provided a comprehensive comparison among DL-based methods for thoracic and head & neck multiorgan segmentation using benchmark datasets, including the 2017 AAPM Thoracic Auto-segmentation Challenge datasets and 2015 MICCAI Head Neck Auto-Segmentation Challenge datasets.
Machine Learning in Quantitative PET Imaging
Jan 18, 2020

Abstract:This paper reviewed the machine learning-based studies for quantitative positron emission tomography (PET). Specifically, we summarized the recent developments of machine learning-based methods in PET attenuation correction and low-count PET reconstruction by listing and comparing the proposed methods, study designs and reported performances of the current published studies with brief discussion on representative studies. The contributions and challenges among the reviewed studies were summarized and highlighted in the discussion part followed by.
Deep Learning in Medical Image Registration: A Review
Dec 27, 2019



Abstract:This paper presents a review of deep learning (DL) based medical image registration methods. We summarized the latest developments and applications of DL-based registration methods in the medical field. These methods were classified into seven categories according to their methods, functions and popularity. A detailed review of each category was presented, highlighting important contributions and identifying specific challenges. A short assessment was presented following the detailed review of each category to summarize its achievements and future potentials. We provided a comprehensive comparison among DL-based methods for lung and brain deformable registration using benchmark datasets. Lastly, we analyzed the statistics of all the cited works from various aspects, revealing the popularity and future trend of development in medical image registration using deep learning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge