Neman Abdoli
Developing a Novel Image Marker to Predict the Responses of Neoadjuvant Chemotherapy for Ovarian Cancer Patients
Sep 13, 2023Abstract:Objective: Neoadjuvant chemotherapy (NACT) is one kind of treatment for advanced stage ovarian cancer patients. However, due to the nature of tumor heterogeneity, the patients' responses to NACT varies significantly among different subgroups. To address this clinical challenge, the purpose of this study is to develop a novel image marker to achieve high accuracy response prediction of the NACT at an early stage. Methods: For this purpose, we first computed a total of 1373 radiomics features to quantify the tumor characteristics, which can be grouped into three categories: geometric, intensity, and texture features. Second, all these features were optimized by principal component analysis algorithm to generate a compact and informative feature cluster. Using this cluster as the input, an SVM based classifier was developed and optimized to create a final marker, indicating the likelihood of the patient being responsive to the NACT treatment. To validate this scheme, a total of 42 ovarian cancer patients were retrospectively collected. A nested leave-one-out cross-validation was adopted for model performance assessment. Results: The results demonstrate that the new method yielded an AUC (area under the ROC [receiver characteristic operation] curve) of 0.745. Meanwhile, the model achieved overall accuracy of 76.2%, positive predictive value of 70%, and negative predictive value of 78.1%. Conclusion: This study provides meaningful information for the development of radiomics based image markers in NACT response prediction.
Evaluating the Effectiveness of 2D and 3D Features for Predicting Tumor Response to Chemotherapy
Apr 14, 2023



Abstract:2D and 3D tumor features are widely used in a variety of medical image analysis tasks. However, for chemotherapy response prediction, the effectiveness between different kinds of 2D and 3D features are not comprehensively assessed, especially in ovarian cancer-related applications. This investigation aims to accomplish such a comprehensive evaluation. For this purpose, CT images were collected retrospectively from 188 advanced-stage ovarian cancer patients. All the metastatic tumors that occurred in each patient were segmented and then processed by a set of six filters. Next, three categories of features, namely geometric, density, and texture features, were calculated from both the filtered results and the original segmented tumors, generating a total of 1595 and 1403 features for the 3D and 2D tumors, respectively. In addition to the conventional single-slice 2D and full-volume 3D tumor features, we also computed the incomplete-3D tumor features, which were achieved by sequentially adding one individual CT slice and calculating the corresponding features. Support vector machine (SVM) based prediction models were developed and optimized for each feature set. 5-fold cross-validation was used to assess the performance of each individual model. The results show that the 2D feature-based model achieved an AUC (area under the ROC curve [receiver operating characteristic]) of 0.84+-0.02. When adding more slices, the AUC first increased to reach the maximum and then gradually decreased to 0.86+-0.02. The maximum AUC was yielded when adding two adjacent slices, with a value of 0.91+-0.01. This initial result provides meaningful information for optimizing machine learning-based decision-making support tools in the future.
Transformers Improve Breast Cancer Diagnosis from Unregistered Multi-View Mammograms
Jun 21, 2022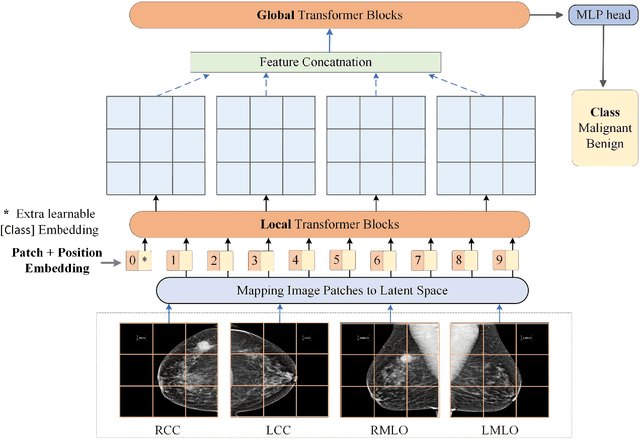
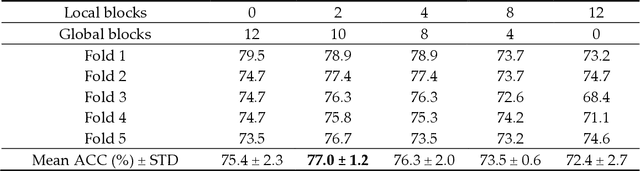
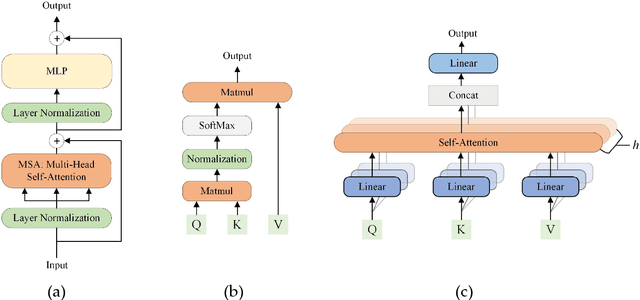
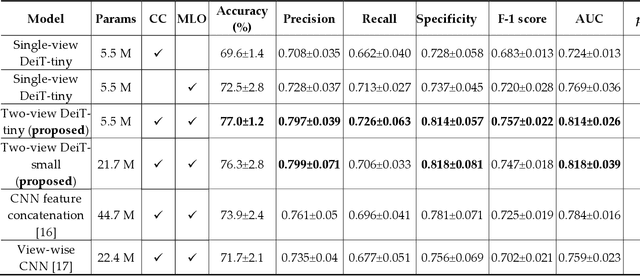
Abstract:Deep convolutional neural networks (CNNs) have been widely used in various medical imaging tasks. However, due to the intrinsic locality of convolution operation, CNNs generally cannot model long-range dependencies well, which are important for accurately identifying or mapping corresponding breast lesion features computed from unregistered multiple mammograms. This motivates us to leverage the architecture of Multi-view Vision Transformers to capture long-range relationships of multiple mammograms from the same patient in one examination. For this purpose, we employ local Transformer blocks to separately learn patch relationships within four mammograms acquired from two-view (CC/MLO) of two-side (right/left) breasts. The outputs from different views and sides are concatenated and fed into global Transformer blocks, to jointly learn patch relationships between four images representing two different views of the left and right breasts. To evaluate the proposed model, we retrospectively assembled a dataset involving 949 sets of mammograms, which include 470 malignant cases and 479 normal or benign cases. We trained and evaluated the model using a five-fold cross-validation method. Without any arduous preprocessing steps (e.g., optimal window cropping, chest wall or pectoral muscle removal, two-view image registration, etc.), our four-image (two-view-two-side) Transformer-based model achieves case classification performance with an area under ROC curve (AUC = 0.818), which significantly outperforms AUC = 0.784 achieved by the state-of-the-art multi-view CNNs (p = 0.009). It also outperforms two one-view-two-side models that achieve AUC of 0.724 (CC view) and 0.769 (MLO view), respectively. The study demonstrates the potential of using Transformers to develop high-performing computer-aided diagnosis schemes that combine four mammograms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge