Yinghui Jiang
LOHA: Direct Graph Spectral Contrastive Learning Between Low-pass and High-pass Views
Jan 06, 2025



Abstract:Spectral Graph Neural Networks effectively handle graphs with different homophily levels, with low-pass filter mining feature smoothness and high-pass filter capturing differences. When these distinct filters could naturally form two opposite views for self-supervised learning, the commonalities between the counterparts for the same node remain unexplored, leading to suboptimal performance. In this paper, a simple yet effective self-supervised contrastive framework, LOHA, is proposed to address this gap. LOHA optimally leverages low-pass and high-pass views by embracing "harmony in diversity". Rather than solely maximizing the difference between these distinct views, which may lead to feature separation, LOHA harmonizes the diversity by treating the propagation of graph signals from both views as a composite feature. Specifically, a novel high-dimensional feature named spectral signal trend is proposed to serve as the basis for the composite feature, which remains relatively unaffected by changing filters and focuses solely on original feature differences. LOHA achieves an average performance improvement of 2.8% over runner-up models on 9 real-world datasets with varying homophily levels. Notably, LOHA even surpasses fully-supervised models on several datasets, which underscores the potential of LOHA in advancing the efficacy of spectral GNNs for diverse graph structures.
GAPS: Geometry-Aware Problem Solver
Jan 29, 2024Abstract:Geometry problem solving presents a formidable challenge within the NLP community. Existing approaches often rely on models designed for solving math word problems, neglecting the unique characteristics of geometry math problems. Additionally, the current research predominantly focuses on geometry calculation problems, while overlooking other essential aspects like proving. In this study, we address these limitations by proposing the Geometry-Aware Problem Solver (GAPS) model. GAPS is specifically designed to generate solution programs for geometry math problems of various types with the help of its unique problem-type classifier. To achieve this, GAPS treats the solution program as a composition of operators and operands, segregating their generation processes. Furthermore, we introduce the geometry elements enhancement method, which enhances the ability of GAPS to recognize geometry elements accurately. By leveraging these improvements, GAPS showcases remarkable performance in resolving geometry math problems. Our experiments conducted on the UniGeo dataset demonstrate the superiority of GAPS over the state-of-the-art model, Geoformer. Specifically, GAPS achieves an accuracy improvement of more than 5.3% for calculation tasks and an impressive 41.1% for proving tasks. Notably, GAPS achieves an impressive accuracy of 97.5% on proving problems, representing a significant advancement in solving geometry proving tasks.
Explaining Graph Neural Networks via Non-parametric Subgraph Matching
Jan 07, 2023



Abstract:The great success in graph neural networks (GNNs) provokes the question about explainability: Which fraction of the input graph is the most determinant of the prediction? Particularly, parametric explainers prevail in existing approaches because of their stronger capability to decipher the black-box (i.e., the target GNN). In this paper, based on the observation that graphs typically share some joint motif patterns, we propose a novel non-parametric subgraph matching framework, dubbed MatchExplainer, to explore explanatory subgraphs. It couples the target graph with other counterpart instances and identifies the most crucial joint substructure by minimizing the node corresponding-based distance. Moreover, we note that present graph sampling or node-dropping methods usually suffer from the false positive sampling problem. To ameliorate that issue, we design a new augmentation paradigm named MatchDrop. It takes advantage of MatchExplainer to fix the most informative portion of the graph and merely operates graph augmentations on the rest less informative part. We conduct extensive experiments on both synthetic and real-world datasets and show the effectiveness of our MatchExplainer by outperforming all parametric baselines with significant margins. Additional results also demonstrate that our MatchDrop is a general scheme to be equipped with GNNs for enhanced performance.
A Score-based Geometric Model for Molecular Dynamics Simulations
Apr 19, 2022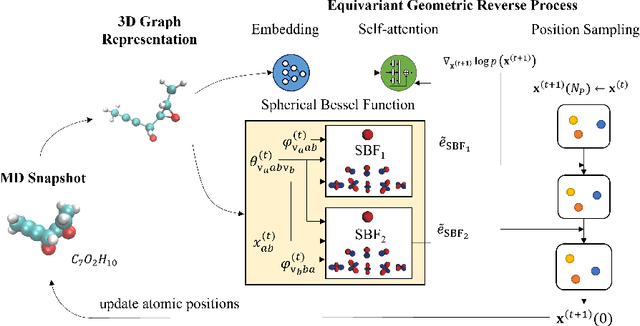
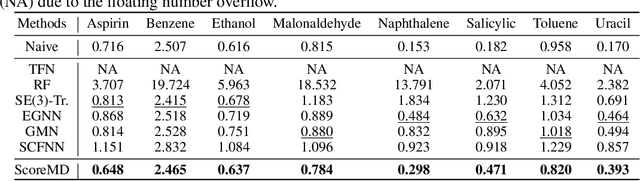
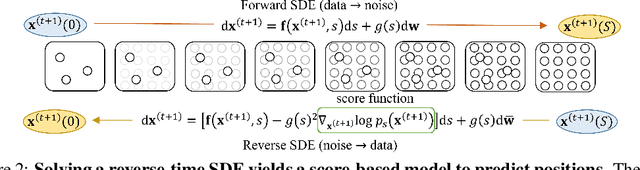

Abstract:Molecular dynamics (MD) has long been the \emph{de facto} choice for modeling complex atomistic systems from first principles, and recently deep learning become a popular way to accelerate it. Notwithstanding, preceding approaches depend on intermediate variables such as the potential energy or force fields to update atomic positions, which requires additional computations to perform back-propagation. To waive this requirement, we propose a novel model called ScoreMD by directly estimating the gradient of the log density of molecular conformations. Moreover, we analyze that diffusion processes highly accord with the principle of enhanced sampling in MD simulations, and is therefore a perfect match to our sequential conformation generation task. That is, ScoreMD perturbs the molecular structure with a conditional noise depending on atomic accelerations and employs conformations at previous timeframes as the prior distribution for sampling. Another challenge of modeling such a conformation generation process is that the molecule is kinetic instead of static, which no prior studies strictly consider. To solve this challenge, we introduce a equivariant geometric Transformer as a score function in the diffusion process to calculate the corresponding gradient. It incorporates the directions and velocities of atomic motions via 3D spherical Fourier-Bessel representations. With multiple architectural improvements, we outperforms state-of-the-art baselines on MD17 and isomers of C7O2H10. This research provides new insights into the acceleration of new material and drug discovery.
Robust Weakly Supervised Learning for COVID-19 Recognition Using Multi-Center CT Images
Dec 09, 2021



Abstract:The world is currently experiencing an ongoing pandemic of an infectious disease named coronavirus disease 2019 (i.e., COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Computed Tomography (CT) plays an important role in assessing the severity of the infection and can also be used to identify those symptomatic and asymptomatic COVID-19 carriers. With a surge of the cumulative number of COVID-19 patients, radiologists are increasingly stressed to examine the CT scans manually. Therefore, an automated 3D CT scan recognition tool is highly in demand since the manual analysis is time-consuming for radiologists and their fatigue can cause possible misjudgment. However, due to various technical specifications of CT scanners located in different hospitals, the appearance of CT images can be significantly different leading to the failure of many automated image recognition approaches. The multi-domain shift problem for the multi-center and multi-scanner studies is therefore nontrivial that is also crucial for a dependable recognition and critical for reproducible and objective diagnosis and prognosis. In this paper, we proposed a COVID-19 CT scan recognition model namely coronavirus information fusion and diagnosis network (CIFD-Net) that can efficiently handle the multi-domain shift problem via a new robust weakly supervised learning paradigm. Our model can resolve the problem of different appearance in CT scan images reliably and efficiently while attaining higher accuracy compared to other state-of-the-art methods.
Weakly Supervised Deep Learning for COVID-19 Infection Detection and Classification from CT Images
Apr 14, 2020



Abstract:An outbreak of a novel coronavirus disease (i.e., COVID-19) has been recorded in Wuhan, China since late December 2019, which subsequently became pandemic around the world. Although COVID-19 is an acutely treated disease, it can also be fatal with a risk of fatality of 4.03% in China and the highest of 13.04% in Algeria and 12.67% Italy (as of 8th April 2020). The onset of serious illness may result in death as a consequence of substantial alveolar damage and progressive respiratory failure. Although laboratory testing, e.g., using reverse transcription polymerase chain reaction (RT-PCR), is the golden standard for clinical diagnosis, the tests may produce false negatives. Moreover, under the pandemic situation, shortage of RT-PCR testing resources may also delay the following clinical decision and treatment. Under such circumstances, chest CT imaging has become a valuable tool for both diagnosis and prognosis of COVID-19 patients. In this study, we propose a weakly supervised deep learning strategy for detecting and classifying COVID-19 infection from CT images. The proposed method can minimise the requirements of manual labelling of CT images but still be able to obtain accurate infection detection and distinguish COVID-19 from non-COVID-19 cases. Based on the promising results obtained qualitatively and quantitatively, we can envisage a wide deployment of our developed technique in large-scale clinical studies.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge